Cell Wall Glycans Mediate Recognition of the Dairy Bacterium Streptococcus thermophilus by Bacteriophages
- PMID: 30242010
- PMCID: PMC6238053
- DOI: 10.1128/AEM.01847-18
Cell Wall Glycans Mediate Recognition of the Dairy Bacterium Streptococcus thermophilus by Bacteriophages
Abstract
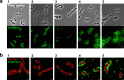
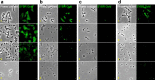
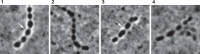
Receptors on the cell surfaces of bacterial hosts are essential during the infection cycle of bacteriophages. To date, the phage receptors of the industrial relevant dairy starter bacterium Streptococcus thermophilus remain elusive. Thus, we set out to identify cell surface structures that are involved in host recognition by dairy streptococcal phages. Five industrial S. thermophilus strains sensitive to different phages (pac type, cos type, and the new type 987), were selected to generate spontaneous bacteriophage-insensitive mutants (BIMs). Of these, approximately 50% were deselected as clustered regularly interspaced short palindromic repeat (CRISPR) mutants, while the other pool was further characterized to identify receptor mutants. On the basis of genome sequencing data, phage resistance in putative receptor mutants was attributed to nucleotide changes in genes encoding glycan biosynthetic pathways. Superresolution structured illumination microscopy was used to visualize the interactions between S. thermophilus and its phages. The phages were either regularly distributed along the cells or located at division sites of the cells. The cell wall structures mediating the latter type of phage adherence were further analyzed via phenotypic and biochemical assays. Altogether, our data suggested that phage adsorption to S. thermophilus is mediated by glycans associated with the bacterial cell surface. Specifically, the pac-type phage CHPC951 adsorbed to polysaccharides anchored to peptidoglycan, while the 987-type phage CHPC926 recognized exocellular polysaccharides associated with the cell surface.IMPORTANCEStreptococcus thermophilus is widely used in starter cultures for cheese and yoghurt production. During dairy fermentations, infections of bacteria with bacteriophages result in acidification failures and a lower quality of the final products. An understanding of the molecular factors involved in phage-host interactions, in particular, the phage receptors in dairy bacteria, is a crucial step for developing better strategies to prevent phage infections in dairy plants.
Keywords: Streptococcus thermophilus; adsorption; bacteriophages; cell wall; glycans; polysaccharides; receptors.
Copyright © 2018 Szymczak et al.
Figures



Similar articles
-
Brussowvirus SW13 Requires a Cell Surface-Associated Polysaccharide To Recognize Its Streptococcus thermophilus Host.Appl Environ Microbiol. 2022 Jan 11;88(1):e0172321. doi: 10.1128/AEM.01723-21. Epub 2021 Oct 20. Appl Environ Microbiol. 2022. PMID: 34669424 Free PMC article.
-
Novel Variants of Streptococcus thermophilus Bacteriophages Are Indicative of Genetic Recombination among Phages from Different Bacterial Species.Appl Environ Microbiol. 2017 Feb 15;83(5):e02748-16. doi: 10.1128/AEM.02748-16. Print 2017 Mar 1. Appl Environ Microbiol. 2017. PMID: 28039135 Free PMC article.
-
A Decade of Streptococcus thermophilus Phage Evolution in an Irish Dairy Plant.Appl Environ Microbiol. 2018 May 1;84(10):e02855-17. doi: 10.1128/AEM.02855-17. Print 2018 May 15. Appl Environ Microbiol. 2018. PMID: 29523549 Free PMC article.
-
Bacteriophage-host interactions in Streptococcus thermophilus and their impact on co-evolutionary processes.FEMS Microbiol Rev. 2023 Jul 5;47(4):fuad032. doi: 10.1093/femsre/fuad032. FEMS Microbiol Rev. 2023. PMID: 37339909 Free PMC article. Review.
-
Dairy lactococcal and streptococcal phage-host interactions: an industrial perspective in an evolving phage landscape.FEMS Microbiol Rev. 2020 Nov 24;44(6):909-932. doi: 10.1093/femsre/fuaa048. FEMS Microbiol Rev. 2020. PMID: 33016324 Review.
Cited by
-
Brussowvirus SW13 Requires a Cell Surface-Associated Polysaccharide To Recognize Its Streptococcus thermophilus Host.Appl Environ Microbiol. 2022 Jan 11;88(1):e0172321. doi: 10.1128/AEM.01723-21. Epub 2021 Oct 20. Appl Environ Microbiol. 2022. PMID: 34669424 Free PMC article.
-
Host-encoded, cell surface-associated exopolysaccharide required for adsorption and infection by lactococcal P335 phage subtypes.Front Microbiol. 2022 Oct 4;13:971166. doi: 10.3389/fmicb.2022.971166. eCollection 2022. Front Microbiol. 2022. PMID: 36267184 Free PMC article.
-
A comparative genomics approach for identifying host-range determinants in Streptococcus thermophilus bacteriophages.Sci Rep. 2019 May 29;9(1):7991. doi: 10.1038/s41598-019-44481-z. Sci Rep. 2019. PMID: 31142793 Free PMC article.
-
Bacteriophage-host interactions as a platform to establish the role of phages in modulating the microbial composition of fermented foods.Microbiome Res Rep. 2022 Jan 12;1(1):3. doi: 10.20517/mrr.2021.04. eCollection 2022. Microbiome Res Rep. 2022. PMID: 38089066 Free PMC article. Review.
-
Fitness Trade-Offs Resulting from Bacteriophage Resistance Potentiate Synergistic Antibacterial Strategies.Infect Immun. 2020 Jun 22;88(7):e00926-19. doi: 10.1128/IAI.00926-19. Print 2020 Jun 22. Infect Immun. 2020. PMID: 32094257 Free PMC article. Review.
References
-
- Lahtinen S, Ouwehand AC, Salminen S, Von Wright A. 2012. Lactic acid bacteria: microbiological and functional aspects, 4th ed CRC Press, Boca Raton, FL.
-
- Binetti AG, Quiberoni A, Reinheimer JA. 2002. Phage adsorption to Streptococcus thermophilus. Influence of environmental factors and characterization of cell-receptors. Food Res Int 35:73–83. doi:10.1016/S0963-9969(01)00121-1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

