ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins
- PMID: 30242134
- PMCID: PMC6187130
- DOI: 10.1073/pnas.1809382115
ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins
Abstract
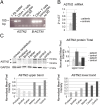
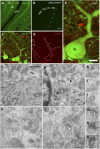
Surface protein dynamics dictate synaptic connectivity and function in neuronal circuits. ASTN2, a gene disrupted by copy number variations (CNVs) in neurodevelopmental disorders, including autism spectrum, was previously shown to regulate the surface expression of ASTN1 in glial-guided neuronal migration. Here, we demonstrate that ASTN2 binds to and regulates the surface expression of multiple synaptic proteins in postmigratory neurons by endocytosis, resulting in modulation of synaptic activity. In cerebellar Purkinje cells (PCs), by immunogold electron microscopy, ASTN2 localizes primarily to endocytic and autophagocytic vesicles in the cell soma and in subsets of dendritic spines. Overexpression of ASTN2 in PCs, but not of ASTN2 lacking the FNIII domain, recurrently disrupted by CNVs in patients, including in a family presented here, increases inhibitory and excitatory postsynaptic activity and reduces levels of ASTN2 binding partners. Our data suggest a fundamental role for ASTN2 in dynamic regulation of surface proteins by endocytic trafficking and protein degradation.
Keywords: autism spectrum disorder; cerebellum; protein degradation; protein trafficking; synapse.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Stitt TN, Hatten ME. Antibodies that recognize astrotactin block granule neuron binding to astroglia. Neuron. 1990;5:639–649. - PubMed
-
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. - PubMed
-
- Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113:755–765. - PubMed
-
- Lesch KP, et al. Molecular genetics of adult ADHD: Converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm (Vienna) 2008;115:1573–1585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

