Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma
- PMID: 30242167
- PMCID: PMC6155108
- DOI: 10.1038/s41598-018-31170-6
Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma
Abstract
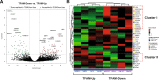
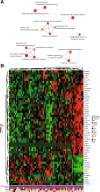
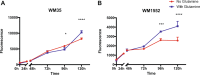
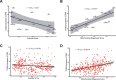
Mitochondria are central key players in cell metabolism, and mitochondrial DNA (mtDNA) instability has been linked to metabolic changes that contribute to tumorigenesis and to increased expression of pro-tumorigenic genes. Here, we use melanoma cell lines and metastatic melanoma tumors to evaluate the effect of mtDNA alterations and the expression of the mtDNA packaging factor, TFAM, on energetic metabolism and pro-tumorigenic nuclear gene expression changes. We report a positive correlation between mtDNA copy number, glucose consumption, and ATP production in melanoma cell lines. Gene expression analysis reveals a down-regulation of glycolytic enzymes in cell lines and an up-regulation of amino acid metabolism enzymes in melanoma tumors, suggesting that TFAM may shift melanoma fuel utilization from glycolysis towards amino acid metabolism, especially glutamine. Indeed, proliferation assays reveal that TFAM-down melanoma cell lines display a growth arrest in glutamine-free media, emphasizing that these cells rely more on glutamine metabolism than glycolysis. Finally, our data indicate that TFAM correlates to VEGF expression and may contribute to tumorigenesis by triggering a more invasive gene expression signature. Our findings contribute to the understanding of how TFAM affects melanoma cell metabolism, and they provide new insight into the mechanisms by which TFAM and mtDNA copy number influence melanoma tumorigenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- 2013/25119-0/Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation)/International
- 2015/21940-7/Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation)/International
- 2013/08135-2/Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation)/International
- 2013/25119-0/Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation)/International
- 2015/21940-7/Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation)/International
- RP170734/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)/International
- 465539/2014-9/Ministry of Science, Technology and Innovation | Conselho Nacional de Desenvolvimento Científico e Tecnológico (National Council for Scientific and Technological Development)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

