PTMD: A Database of Human Disease-associated Post-translational Modifications
- PMID: 30244175
- PMCID: PMC6205080
- DOI: 10.1016/j.gpb.2018.06.004
PTMD: A Database of Human Disease-associated Post-translational Modifications
Abstract
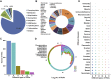
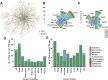
Various posttranslational modifications (PTMs) participate in nearly all aspects of biological processes by regulating protein functions, and aberrant states of PTMs are frequently implicated in human diseases. Therefore, an integral resource of PTM-disease associations (PDAs) would be a great help for both academic research and clinical use. In this work, we reported PTMD, a well-curated database containing PTMs that are associated with human diseases. We manually collected 1950 known PDAs in 749 proteins for 23 types of PTMs and 275 types of diseases from the literature. Database analyses show that phosphorylation has the largest number of disease associations, whereas neurologic diseases have the largest number of PTM associations. We classified all known PDAs into six classes according to the PTM status in diseases and demonstrated that the upregulation and presence of PTM events account for a predominant proportion of disease-associated PTM events. By reconstructing a disease-gene network, we observed that breast cancers have the largest number of associated PTMs and AKT1 has the largest number of PTMs connected to diseases. Finally, the PTMD database was developed with detailed annotations and can be a useful resource for further analyzing the relations between PTMs and human diseases. PTMD is freely accessible at http://ptmd.biocuckoo.org.
Keywords: AKT1; Disease–gene network; PTM–disease association; Phosphorylation; Posttranslational modification.
Copyright © 2018 The Authors. Production and hosting by Elsevier B.V. All rights reserved.
Figures


References
-
- Mann M., Jensen O.N. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255. - PubMed
-
- Hendriks I.A., Lyon D., Young C., Jensen L.J., Vertegaal A.C., Nielsen M.L. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325–336. - PubMed
-
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

