Mode of Action of Kanglemycin A, an Ansamycin Natural Product that Is Active against Rifampicin-Resistant Mycobacterium tuberculosis
- PMID: 30244835
- PMCID: PMC6202310
- DOI: 10.1016/j.molcel.2018.08.028
Mode of Action of Kanglemycin A, an Ansamycin Natural Product that Is Active against Rifampicin-Resistant Mycobacterium tuberculosis
Abstract
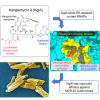
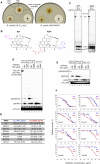
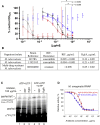
Antibiotic-resistant bacterial pathogens pose an urgent healthcare threat, prompting a demand for new medicines. We report the mode of action of the natural ansamycin antibiotic kanglemycin A (KglA). KglA binds bacterial RNA polymerase at the rifampicin-binding pocket but maintains potency against RNA polymerases containing rifampicin-resistant mutations. KglA has antibiotic activity against rifampicin-resistant Gram-positive bacteria and multidrug-resistant Mycobacterium tuberculosis (MDR-M. tuberculosis). The X-ray crystal structures of KglA with the Escherichia coli RNA polymerase holoenzyme and Thermus thermophilus RNA polymerase-promoter complex reveal an altered-compared with rifampicin-conformation of KglA within the rifampicin-binding pocket. Unique deoxysugar and succinate ansa bridge substituents make additional contacts with a separate, hydrophobic pocket of RNA polymerase and preclude the formation of initial dinucleotides, respectively. Previous ansa-chain modifications in the rifamycin series have proven unsuccessful. Thus, KglA represents a key starting point for the development of a new class of ansa-chain derivatized ansamycins to tackle rifampicin resistance.
Keywords: MDR-TB; RNA polymerase; antibiotic resistance; antibiotics; holo-enzyme crystal structure; kanglemycin A; multidrug-resistant Mycobacterium tuberculosis; promoter complex crystal structure; rifampicin; rifamycin.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Andrews J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48(Suppl 1):5–16. - PubMed
-
- Aristoff P.A., Garcia G.A., Kirchhoff P.D., Showalter H.D. Rifamycins--obstacles and opportunities. Tuberculosis (Edinb.) 2010;90:94–118. - PubMed
-
- Artsimovitch I., Vassylyeva M.N., Svetlov D., Svetlov V., Perederina A., Igarashi N., Matsugaki N., Wakatsuki S., Tahirov T.H., Vassylyev D.G. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

