Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer
- PMID: 30244975
- PMCID: PMC6224605
- DOI: 10.1016/j.cmet.2018.08.021
Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer
Abstract
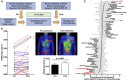
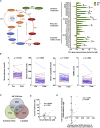
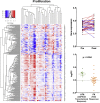
Late-phase clinical trials investigating metformin as a cancer therapy are underway. However, there remains controversy as to the mode of action of metformin in tumors at clinical doses. We conducted a clinical study integrating measurement of markers of systemic metabolism, dynamic FDG-PET-CT, transcriptomics, and metabolomics at paired time points to profile the bioactivity of metformin in primary breast cancer. We show metformin reduces the levels of mitochondrial metabolites, activates multiple mitochondrial metabolic pathways, and increases 18-FDG flux in tumors. Two tumor groups are identified with distinct metabolic responses, an OXPHOS transcriptional response (OTR) group for which there is an increase in OXPHOS gene transcription and an FDG response group with increased 18-FDG uptake. Increase in proliferation, as measured by a validated proliferation signature, suggested that patients in the OTR group were resistant to metformin treatment. We conclude that mitochondrial response to metformin in primary breast cancer may define anti-tumor effect.
Trial registration: ClinicalTrials.gov NCT01266486.
Keywords: breast neoplasms; cancer metabolism; clinical study; gene expression profiling; metabolomics; metformin; mitochondria; positron emission tomography.
Crown Copyright © 2018. Published by Elsevier Inc. All rights reserved.
Figures

Comment in
-
In search of the mechanisms of metformin in cancer.Nat Rev Endocrinol. 2018 Nov;14(11):628. doi: 10.1038/s41574-018-0104-3. Nat Rev Endocrinol. 2018. PMID: 30258089 No abstract available.
References
-
- Ben Sahra I., Laurent K., Giuliano S., Larbret F., Ponzio G., Gounon P., Le Marchand-Brustel Y., Giorgetti-Peraldi S., Cormont M., Bertolotto C. Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res. 2010;70:2465–2475. - PubMed
-
- Bertoldo A., Peltoniemi P., Oikonen V., Knuuti J., Nuutila P., Cobelli C. Kinetic modeling of [(18)F]FDG in skeletal muscle by PET: a four-compartment five-rate-constant model. Am. J. Physiol. Endocrinol. Metab. 2001;281:E524–E536. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

