KAT6A Syndrome: genotype-phenotype correlation in 76 patients with pathogenic KAT6A variants
- PMID: 30245513
- PMCID: PMC6634310
- DOI: 10.1038/s41436-018-0259-2
KAT6A Syndrome: genotype-phenotype correlation in 76 patients with pathogenic KAT6A variants
Erratum in
-
Correction: KAT6A Syndrome: genotype-phenotype correlation in 76 patients with pathogenic KAT6A variants.Genet Med. 2020 Nov;22(11):1920. doi: 10.1038/s41436-020-00944-7. Genet Med. 2020. PMID: 32814847
Abstract
Purpose: Pathogenic variants in KAT6A have recently been identified as a cause of syndromic developmental delay. Within 2 years, the number of patients identified with pathogenic KAT6A variants has rapidly expanded and the full extent and variability of the clinical phenotype has not been reported.
Methods: We obtained data for patients with KAT6A pathogenic variants through three sources: treating clinicians, an online family survey distributed through social media, and a literature review.
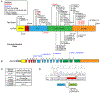
Results: We identified 52 unreported cases, bringing the total number of published cases to 76. Our results expand the genotypic spectrum of pathogenic variants to include missense and splicing mutations. We functionally validated a pathogenic splice-site variant and identified a likely hotspot location for de novo missense variants. The majority of clinical features in KAT6A syndrome have highly variable penetrance. For core features such as intellectual disability, speech delay, microcephaly, cardiac anomalies, and gastrointestinal complications, genotype- phenotype correlations show that late-truncating pathogenic variants (exons 16-17) are significantly more prevalent. We highlight novel associations, including an increased risk of gastrointestinal obstruction.
Conclusion: Our data expand the genotypic and phenotypic spectrum for individuals with genetic pathogenic variants in KAT6A and we outline appropriate clinical management.
Keywords: KAT6A syndrome; chromatin modifiers; intellectual disability; genetic diagnosis; phenotypic spectrum.
Figures


References
-
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26(37):5395–5407. - PubMed
-
- Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell. 2009;17(5):674–686. - PubMed
-
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, Fox M, Fogel BL, Martinez-Agosto JA, Wong DA, Chang VY, Shieh PB, Palmer CGS, Dipple KM, Grody WW, Vilain E, Nelson SF. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. Journal of American Medical Association 2014(in press). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

