DNA methylation biomarkers for hepatocellular carcinoma
- PMID: 30245591
- PMCID: PMC6142709
- DOI: 10.1186/s12935-018-0629-5
DNA methylation biomarkers for hepatocellular carcinoma
Abstract
Background: Aberrant methylation of DNA is a key driver of hepatocellular carcinoma (HCC). In this study, we sought to integrate four cohorts profile datasets to identify such abnormally methylated genes and pathways associated with HCC.
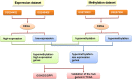
Methods: To this end, we downloaded microarray datasets examining gene expression (GSE84402, GSE46408) and gene methylation (GSE73003, GSE57956) from the GEO database. Abnormally methylated differentially expressed genes (DEGs) were sorted and pathways were analyzed. The String database was then used to perform enrichment and functional analysis of identified pathways and genes. Cytoscape software was used to create a protein-protein interaction network, and MCODE was used for module analysis. Finally, overall survival analysis of hub genes was performed by the OncoLnc online tool.
Results: In total, we identified 19 hypomethylated highly expressed genes and 14 hypermethylated lowly expressed genes at the screening step, and finally found six mostly changed hub genes including MAD2L1, CDC20, CCNB1, CCND1, AR and ESR1. Pathway analysis showed that aberrantly methylated-DEGs mainly associated with the cell cycle process, p53 signaling, and MAPK signaling in HCC. After validation in TCGA database, the methylation and expression status of hub genes was significantly altered and same with our results. Patients with high expression of MAD2L1, CDC20 and CCNB1 and low expression of CCND1, AR, and ESR1 was associated with shorter overall survival.
Conclusions: Taken together, we have identified novel aberrantly methylated genes and pathways linked to HCC, potentially offering novel insights into the molecular mechanisms governing HCC progression and serving as novel biomarkers for precision diagnosis and disease treatment.
Keywords: Hepatocellular carcinoma; Hub genes; Methylation.
Figures






References
-
- Addeo R, Caraglia M, Del Prete S. Highlights of regional meeting of Italian Southern Oncological Group (GOIM): focus on hepatocellular carcinoma: biological and clinical background, therapeutic guide-lines and perspectives. 7 November 2008, Naples, Italy. Expert Opin Investig Drugs. 2009;18(3):373–378. doi: 10.1517/14712590802680158. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

