Comprehensive analysis of the BC200 ribonucleoprotein reveals a reciprocal regulatory function with CSDE1/UNR
- PMID: 30247708
- PMCID: PMC6265466
- DOI: 10.1093/nar/gky860
Comprehensive analysis of the BC200 ribonucleoprotein reveals a reciprocal regulatory function with CSDE1/UNR
Abstract
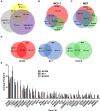
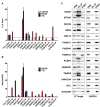
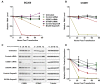
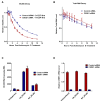
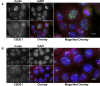
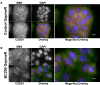
BC200 is a long non-coding RNA primarily expressed in brain but aberrantly expressed in various cancers. To gain a further understanding of the function of BC200, we performed proteomic analyses of the BC200 ribonucleoprotein (RNP) by transfection of 3' DIG-labelled BC200. Protein binding partners of the functionally related murine RNA BC1 as well as a scrambled BC200 RNA were also assessed in both human and mouse cell lines. Stringent validation of proteins identified by mass spectrometry confirmed 14 of 84 protein binding partners and excluded eight proteins that did not appreciably bind BC200 in reverse experiments. Gene ontology analyses revealed general roles in RNA metabolic processes, RNA processing and splicing. Protein/RNA interaction sites were mapped with a series of RNA truncations revealing three distinct modes of interaction involving either the 5' Alu-domain, 3' A-rich or 3' C-rich regions. Due to their high enrichment values in reverse experiments, CSDE1 and STRAP were further analyzed demonstrating a direct interaction between CSDE1 and BC200 and indirect binding of STRAP to BC200 via heterodimerization with CSDE1. Knock-down studies identified a reciprocal regulatory relationship between CSDE1 and BC200 and immunofluorescence analysis of BC200 knock-down cells demonstrated a dramatic reorganization of CSDE1 into distinct nuclear foci.
Figures





Similar articles
-
The noncoding RNA BC200 associates with polysomes to positively regulate mRNA translation in tumor cells.J Biol Chem. 2021 Jan-Jun;296:100036. doi: 10.1074/jbc.RA120.015775. Epub 2020 Nov 24. J Biol Chem. 2021. PMID: 33410401 Free PMC article.
-
Identification of human autoantigen La/SS-B as BC1/BC200 RNA-binding protein.DNA Cell Biol. 1998 Sep;17(9):751-9. doi: 10.1089/dna.1998.17.751. DNA Cell Biol. 1998. PMID: 9778034
-
The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation.Mol Cancer. 2017 Jun 26;16(1):109. doi: 10.1186/s12943-017-0679-7. Mol Cancer. 2017. PMID: 28651607 Free PMC article.
-
The role of CSDE1 in translational reprogramming and human diseases.Cell Commun Signal. 2020 Jan 27;18(1):14. doi: 10.1186/s12964-019-0496-2. Cell Commun Signal. 2020. PMID: 31987048 Free PMC article. Review.
-
Regulation of the expression and activity of Unr in mammalian cells.Biochem Soc Trans. 2015 Dec;43(6):1241-6. doi: 10.1042/BST20150165. Biochem Soc Trans. 2015. PMID: 26614667 Review.
Cited by
-
An RNA guanine quadruplex regulated pathway to TRAIL-sensitization by DDX21.RNA. 2020 Jan;26(1):44-57. doi: 10.1261/rna.072199.119. Epub 2019 Oct 25. RNA. 2020. PMID: 31653714 Free PMC article.
-
RNA Polymerase III Subunit Mutations in Genetic Diseases.Front Mol Biosci. 2021 Jul 30;8:696438. doi: 10.3389/fmolb.2021.696438. eCollection 2021. Front Mol Biosci. 2021. PMID: 34395528 Free PMC article. Review.
-
Brain Cytoplasmic RNAs in Neurons: From Biosynthesis to Function.Biomolecules. 2020 Feb 17;10(2):313. doi: 10.3390/biom10020313. Biomolecules. 2020. PMID: 32079202 Free PMC article. Review.
-
Brain Long Noncoding RNAs: Multitask Regulators of Neuronal Differentiation and Function.Molecules. 2021 Jun 28;26(13):3951. doi: 10.3390/molecules26133951. Molecules. 2021. PMID: 34203457 Free PMC article. Review.
-
Nuclear SRP9/SRP14 heterodimer transcriptionally regulates 7SL and BC200 RNA expression.RNA. 2023 Aug;29(8):1185-1200. doi: 10.1261/rna.079649.123. Epub 2023 May 8. RNA. 2023. PMID: 37156570 Free PMC article.
References
-
- Chen W., Bocker W., Brosius J., Tiedge H.. Expression of neural BC200 RNA in human tumours. J. Pathol. 1997; 183:345–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

