Evolutionary emergence of infectious diseases in heterogeneous host populations
- PMID: 30248089
- PMCID: PMC6171948
- DOI: 10.1371/journal.pbio.2006738
Evolutionary emergence of infectious diseases in heterogeneous host populations
Abstract
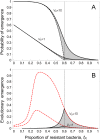
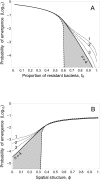
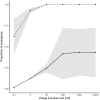
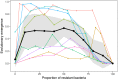
The emergence and re-emergence of pathogens remains a major public health concern. Unfortunately, when and where pathogens will (re-)emerge is notoriously difficult to predict, as the erratic nature of those events is reinforced by the stochastic nature of pathogen evolution during the early phase of an epidemic. For instance, mutations allowing pathogens to escape host resistance may boost pathogen spread and promote emergence. Yet, the ecological factors that govern such evolutionary emergence remain elusive because of the lack of ecological realism of current theoretical frameworks and the difficulty of experimentally testing their predictions. Here, we develop a theoretical model to explore the effects of the heterogeneity of the host population on the probability of pathogen emergence, with or without pathogen evolution. We show that evolutionary emergence and the spread of escape mutations in the pathogen population is more likely to occur when the host population contains an intermediate proportion of resistant hosts. We also show that the probability of pathogen emergence rapidly declines with the diversity of resistance in the host population. Experimental tests using lytic bacteriophages infecting their bacterial hosts containing Clustered Regularly Interspaced Short Palindromic Repeat and CRISPR-associated (CRISPR-Cas) immune defenses confirm these theoretical predictions. These results suggest effective strategies for cross-species spillover and for the management of emerging infectious diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
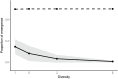
Figures
References
-
- Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Wiley Online Library; 1992.
-
- Diekmann O, Heesterbeek H, Britton T. Mathematical tools for understanding infectious disease dynamics. Princeton University Press; 2013.
-
- Keeling MJ, Rohani P. Modeling infectious diseases in humans and animals. Princeton: Princeton University Press; 2008.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

