Analysis of Drosophila melanogaster testis transcriptome
- PMID: 30249207
- PMCID: PMC6154878
- DOI: 10.1186/s12864-018-5085-z
Analysis of Drosophila melanogaster testis transcriptome
Abstract
Background: The formation of matured and individual sperm involves a series of molecular and spectacular morphological changes of the developing cysts in Drosophila melanogaster testis. Recent advances in RNA Sequencing (RNA-Seq) technology help us to understand the complexity of eukaryotic transcriptomes by dissecting different tissues and developmental stages of organisms. To gain a better understanding of cellular differentiation of spermatogenesis, we applied RNA-Seq to analyse the testis-specific transcriptome, including coding and non-coding genes.
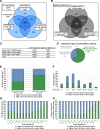
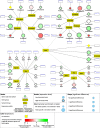
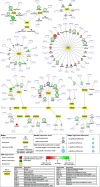
Results: We isolated three different parts of the wild-type testis by dissecting and cutting the different regions: 1.) the apical region, which contains stem cells and developing spermatocytes 2.) the middle region, with enrichment of meiotic cysts 3.) the basal region, which contains elongated post-meiotic cysts with spermatids. Total RNA was isolated from each region and analysed by next-generation sequencing. We collected data from the annotated 17412 Drosophila genes and identified 5381 genes with significant transcript accumulation differences between the regions, representing the main stages of spermatogenesis. We demonstrated for the first time the presence and region specific distribution of 2061 lncRNAs in testis, with 203 significant differences. Using the available modENCODE RNA-Seq data, we determined the tissue specificity indices of Drosophila genes. Combining the indices with our results, we identified genes with region-specific enrichment in testis.
Conclusion: By multiple analyses of our results and integrating existing knowledge about Drosophila melanogaster spermatogenesis to our dataset, we were able to describe transcript composition of different regions of Drosophila testis, including several stage-specific transcripts. We present searchable visualizations that can facilitate the identification of new components that play role in the organisation and composition of different stages of spermatogenesis, including the less known, but complex regulation of post-meiotic stages.
Keywords: Drosophila; RNA sequencing; Spermatogenesis; Testis; Transcriptome.
Conflict of interest statement
Ethics approval and consent to participate
This study does not involve human samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





Similar articles
-
Genome-wide identification and developmental expression profiling of long noncoding RNAs during Drosophila metamorphosis.Sci Rep. 2016 Mar 21;6:23330. doi: 10.1038/srep23330. Sci Rep. 2016. PMID: 26996731 Free PMC article.
-
RNA profiling of human testicular cells identifies syntenic lncRNAs associated with spermatogenesis.Hum Reprod. 2019 Jul 8;34(7):1278-1290. doi: 10.1093/humrep/dez063. Hum Reprod. 2019. PMID: 31247106
-
Isoform-resolution single-cell RNA sequencing reveals the transcriptional panorama of adult Baoshan pig testis cells.BMC Genomics. 2025 May 8;26(1):459. doi: 10.1186/s12864-025-11636-4. BMC Genomics. 2025. PMID: 40340725 Free PMC article.
-
Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies.Reproduction. 2014 Apr 8;147(5):R131-41. doi: 10.1530/REP-13-0594. Print 2014 May. Reproduction. 2014. PMID: 24713396 Review.
-
Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis.Heredity (Edinb). 2009 Jul;103(1):23-31. doi: 10.1038/hdy.2009.23. Epub 2009 Mar 11. Heredity (Edinb). 2009. PMID: 19277057 Review.
Cited by
-
A comparative analysis of fruit fly and human glutamate dehydrogenases in Drosophila melanogaster sperm development.Front Cell Dev Biol. 2023 Nov 2;11:1281487. doi: 10.3389/fcell.2023.1281487. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38020911 Free PMC article.
-
The tumor suppressor archipelago E3 ligase is required for spermatid differentiation in Drosophila testis.Sci Rep. 2021 Apr 19;11(1):8422. doi: 10.1038/s41598-021-87656-3. Sci Rep. 2021. PMID: 33875704 Free PMC article.
-
Diallyl Trisulfide Causes Male Infertility with Oligoasthenoteratospermia in Sitotroga cerealella through the Ubiquitin-Proteasome Pathway.Cells. 2023 Oct 23;12(20):2507. doi: 10.3390/cells12202507. Cells. 2023. PMID: 37887351 Free PMC article.
-
An orphan gene is essential for efficient sperm entry into eggs in Drosophila melanogaster.bioRxiv [Preprint]. 2024 Dec 14:2024.08.08.607187. doi: 10.1101/2024.08.08.607187. bioRxiv. 2024. Update in: Genetics. 2025 Mar 17;229(3):iyaf008. doi: 10.1093/genetics/iyaf008. PMID: 39149251 Free PMC article. Updated. Preprint.
-
An orphan gene is essential for efficient sperm entry into eggs in Drosophila melanogaster.Genetics. 2025 Mar 17;229(3):iyaf008. doi: 10.1093/genetics/iyaf008. Genetics. 2025. PMID: 39903197
References
-
- Fuller MT. Spermatogenesis. In: B M, MA A, editors. Dev Drosoph melanogaster. New York, US: COLD SPRING HARBOR LABORATORY PRESS; 1993. pp. 71–148.
-
- Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125:1833–1843. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

