Pleiotropic Impacts of Macrophage and Microglial Deficiency on Development in Rats with Targeted Mutation of the Csf1r Locus
- PMID: 30249809
- PMCID: PMC6196293
- DOI: 10.4049/jimmunol.1701783
Pleiotropic Impacts of Macrophage and Microglial Deficiency on Development in Rats with Targeted Mutation of the Csf1r Locus
Erratum in
-
Correction: Pleiotropic Impacts of Macrophage and Microglial Deficiency on Development in Rats with Targeted Mutation of the Csf1r Locus.J Immunol. 2019 Jun 1;202(11):3334-3335. doi: 10.4049/jimmunol.1900420. Epub 2019 Apr 26. J Immunol. 2019. PMID: 31028122 Free PMC article. No abstract available.
Abstract
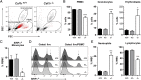
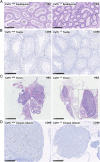
We have produced Csf1r-deficient rats by homologous recombination in embryonic stem cells. Consistent with the role of Csf1r in macrophage differentiation, there was a loss of peripheral blood monocytes, microglia in the brain, epidermal Langerhans cells, splenic marginal zone macrophages, bone-associated macrophages and osteoclasts, and peritoneal macrophages. Macrophages of splenic red pulp, liver, lung, and gut were less affected. The pleiotropic impacts of the loss of macrophages on development of multiple organ systems in rats were distinct from those reported in mice. Csf1r-/- rats survived well into adulthood with postnatal growth retardation, distinct skeletal and bone marrow abnormalities, infertility, and loss of visceral adipose tissue. Gene expression analysis in spleen revealed selective loss of transcripts associated with the marginal zone and, in brain regions, the loss of known and candidate novel microglia-associated transcripts. Despite the complete absence of microglia, there was little overt phenotype in brain, aside from reduced myelination and increased expression of dopamine receptor-associated transcripts in striatum. The results highlight the redundant and nonredundant functions of CSF1R signaling and of macrophages in development, organogenesis, and homeostasis.
Copyright © 2018 The Authors.
Figures








References
-
- Okabe Y., Medzhitov R. 2016. Tissue biology perspective on macrophages. Nat. Immunol. 17: 9–17. - PubMed
-
- Garceau V., Smith J., Paton I. R., Davey M., Fares M. A., Sester D. P., Burt D. W., Hume D. A. 2010. Pivotal advance: avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J. Leukoc. Biol. 87: 753–764. - PubMed
-
- Sasmono R. T., Oceandy D., Pollard J. W., Tong W., Pavli P., Wainwright B. J., Ostrowski M. C., Himes S. R., Hume D. A. 2003. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101: 1155–1163. - PubMed
-
- Balic A., Garcia-Morales C., Vervelde L., Gilhooley H., Sherman A., Garceau V., Gutowska M. W., Burt D. W., Kaiser P., Hume D. A., Sang H. M. 2014. Visualisation of chicken macrophages using transgenic reporter genes: insights into the development of the avian macrophage lineage. Development 141: 3255–3265. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

