An Efficient Algorithm for Sensitively Detecting Circular RNA from RNA-seq Data
- PMID: 30250003
- PMCID: PMC6213952
- DOI: 10.3390/ijms19102897
An Efficient Algorithm for Sensitively Detecting Circular RNA from RNA-seq Data
Abstract
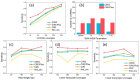
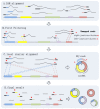
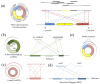
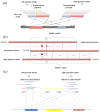
Circular RNA (circRNA) is an important member of non-coding RNA family. Numerous computational methods for detecting circRNAs from RNA-seq data have been developed in the past few years, but there are dramatic differences among the algorithms regarding the balancing of the sensitivity and precision of the detection and filtering strategies. To further improve the sensitivity, while maintaining an acceptable precision of circRNA detection, a novel and efficient de novo detection algorithm, CIRCPlus, is proposed in this paper. CIRCPlus accurately locates circRNA candidates by identifying a set of back-spliced junction reads by comparing the local similar sequence of each pair of spanning junction reads. This strategy, thus, utilizes the important information provided by unbalanced spanning reads, which facilitates the detection especially when the expression levels of circRNA are unapparent. The performance of CIRCPlus was tested and compared to the existing de novo methods on the real datasets as well as a series of simulation datasets with different configurations. The experiment results demonstrated that the sensitivities of CIRCPlus were able to reach 90% in common simulation settings, while CIRCPlus held balanced sensitivity and reliability on the real datasets according to an objective assessment criteria based on RNase R-treated samples. The software tool is available for academic uses only.
Keywords: RNA-seq; de novo detection; high sensitivity; local similar sequence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

