SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis
- PMID: 30250022
- PMCID: PMC6155123
- DOI: 10.1038/s41419-018-0962-6
SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis
Abstract
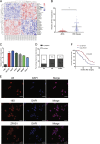
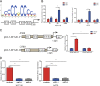
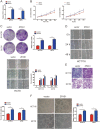
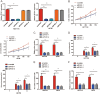
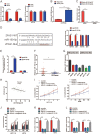
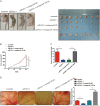
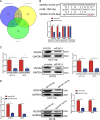
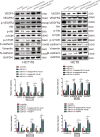
Increasing long non-coding RNAs (lncRNAs) have been reported to play key roles in the development and progression of various malignancies. ZNFX1 antisense RNA1 (ZFAS1) has been reported to be aberrant expression and suggested as a tumor suppressor or oncogene in many cancers. However, the biological role and underlying molecular mechanism of ZFAS1, especially the miRNA sponge role of which in CRC remain largely unknown. We found that ZFAS1 expression was higher in CRC tissues, where it was associated with poor overall survival (OS), we also showed that ZFAS1 upregulation was induced by nuclear transcription factor SP1. Moreover, ZFAS1 and VEGFA are both targets of miR-150-5p, while ZFAS1 binds to miR-150-5p in an AGO2-dependent manner. Additionally, ZFAS1 upregulation markedly promoted as well as ZFAS1 knockdown significantly suppressed CRC cell proliferation, migration, invasion and angiogenesis, and the inhibitory effect caused by ZFAS1 knockdown could be reversed by antagomiR-150-5p. Lastly, we demonstrated that ZFAS1 knockdown inhibited EMT process and inactivated VEGFA/VEGFR2 and downstream Akt/mTOR signaling pathway in CRC. Our data demonstrated that SP1-induced ZFAS1 contributed to CRC progression by upregulating VEGFA via competitively binding to miR-150-5p, which acts as a tumor suppressor by targeting VEGFA in CRC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Long Non-coding RNA Zinc Finger Antisense 1 (ZFAS1) Regulates Proliferation, Migration, Invasion, and Apoptosis by Targeting MiR-7-5p in Colorectal Cancer.Med Sci Monit. 2019 Jul 11;25:5150-5158. doi: 10.12659/MSM.916619. Med Sci Monit. 2019. PMID: 31295229 Free PMC article.
-
P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2.Cell Death Dis. 2019 Feb 12;10(2):131. doi: 10.1038/s41419-018-1188-3. Cell Death Dis. 2019. PMID: 30755600 Free PMC article.
-
Long noncoding RNA ZFAS1 promotes tumorigenesis through regulation of miR-150-5p/RAB9A in melanoma.Melanoma Res. 2019 Dec;29(6):569-581. doi: 10.1097/CMR.0000000000000595. Melanoma Res. 2019. PMID: 30889053
-
Research Progress of Long Non-coding RNA-ZFAS1 in Malignant Tumors.Cell Biochem Biophys. 2024 Dec;82(4):3145-3156. doi: 10.1007/s12013-024-01441-3. Epub 2024 Jul 26. Cell Biochem Biophys. 2024. PMID: 39060915 Review.
-
ZFAS1: A novel vital oncogenic lncRNA in multiple human cancers.Cell Prolif. 2019 Jan;52(1):e12513. doi: 10.1111/cpr.12513. Epub 2018 Oct 5. Cell Prolif. 2019. PMID: 30288832 Free PMC article. Review.
Cited by
-
SP1-Mediated Upregulation of lncRNA LINC01614 Functions a ceRNA for miR-383 to Facilitate Glioma Progression Through Regulation of ADAM12.Onco Targets Ther. 2020 May 18;13:4305-4318. doi: 10.2147/OTT.S242854. eCollection 2020. Onco Targets Ther. 2020. PMID: 32547064 Free PMC article.
-
Angioregulatory microRNAs in Colorectal Cancer.Cancers (Basel). 2019 Dec 26;12(1):71. doi: 10.3390/cancers12010071. Cancers (Basel). 2019. PMID: 31887997 Free PMC article. Review.
-
Inflammatory-Dependent Bidirectional Effect of Bile Acids on NLRP3 Inflammasome and Its Role in Ameliorating CPT-11-Induced Colitis.Front Pharmacol. 2022 May 31;13:677738. doi: 10.3389/fphar.2022.677738. eCollection 2022. Front Pharmacol. 2022. PMID: 35712724 Free PMC article.
-
Long Non-coding RNA Zinc Finger Antisense 1 (ZFAS1) Regulates Proliferation, Migration, Invasion, and Apoptosis by Targeting MiR-7-5p in Colorectal Cancer.Med Sci Monit. 2019 Jul 11;25:5150-5158. doi: 10.12659/MSM.916619. Med Sci Monit. 2019. PMID: 31295229 Free PMC article.
-
Long non-coding RNA ZFAS1 promotes the expression of EPAS1 in gastric cardia adenocarcinoma.J Adv Res. 2020 Jun 24;28:7-15. doi: 10.1016/j.jare.2020.06.006. eCollection 2021 Feb. J Adv Res. 2020. PMID: 33364040 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

