Fam49/CYRI interacts with Rac1 and locally suppresses protrusions
- PMID: 30250061
- PMCID: PMC6863750
- DOI: 10.1038/s41556-018-0198-9
Fam49/CYRI interacts with Rac1 and locally suppresses protrusions
Abstract
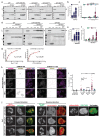
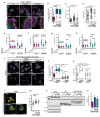
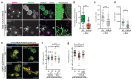
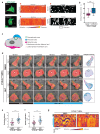
Actin-based protrusions are reinforced through positive feedback, but it is unclear what restricts their size, or limits positive signals when they retract or split. We identify an evolutionarily conserved regulator of actin-based protrusion: CYRI (CYFIP-related Rac interactor) also known as Fam49 (family of unknown function 49). CYRI binds activated Rac1 via a domain of unknown function (DUF1394) shared with CYFIP, defining DUF1394 as a Rac1-binding module. CYRI-depleted cells have broad lamellipodia enriched in Scar/WAVE, but reduced protrusion-retraction dynamics. Pseudopods induced by optogenetic Rac1 activation in CYRI-depleted cells are larger and longer lived. Conversely, CYRI overexpression suppresses recruitment of active Scar/WAVE to the cell edge, resulting in short-lived, unproductive protrusions. CYRI thus focuses protrusion signals and regulates pseudopod complexity by inhibiting Scar/WAVE-induced actin polymerization. It thus behaves like a 'local inhibitor' as predicted in widely accepted mathematical models, but not previously identified in cells. CYRI therefore regulates chemotaxis, cell migration and epithelial polarization by controlling the polarity and plasticity of protrusions.
Figures


References
-
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. - PubMed
-
- Insall R. The interaction between pseudopods and extracellular signalling during chemotaxis and directed migration. Curr Opin Cell Biol. 2013;25:526–531. - PubMed
-
- Davidson AJ, Insall RH. Actin-based motility: WAVE regulatory complex structure reopens old SCARs. Curr Biol. 2011;21:R66–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 24450/CRUK_/Cancer Research UK/United Kingdom
- 12935/CRUK_/Cancer Research UK/United Kingdom
- A15672/CRUK_/Cancer Research UK/United Kingdom
- R01 EY025205/EY/NEI NIH HHS/United States
- A15673/CRUK_/Cancer Research UK/United Kingdom
- 19257/CRUK_/Cancer Research UK/United Kingdom
- A12935/CRUK_/Cancer Research UK/United Kingdom
- A19257/CRUK_/Cancer Research UK/United Kingdom
- BB/L022087/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 20017/CRUK_/Cancer Research UK/United Kingdom
- 24452/CRUK_/Cancer Research UK/United Kingdom
- 29800/CRUK_/Cancer Research UK/United Kingdom
- 15565/CRUK_/Cancer Research UK/United Kingdom
- MR/R011842/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

