Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements
- PMID: 30250208
- PMCID: PMC6155145
- DOI: 10.1038/s41467-018-06393-w
Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements
Abstract
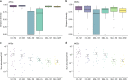
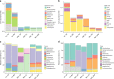
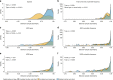
The infant gut microbiota has a high abundance of antibiotic resistance genes (ARGs) compared to adults, even in the absence of antibiotic exposure. Here we study potential sources of infant gut ARGs by performing metagenomic sequencing of breast milk, as well as infant and maternal gut microbiomes. We find that fecal ARG and mobile genetic element (MGE) profiles of infants are more similar to those of their own mothers than to those of unrelated mothers. MGEs in mothers' breast milk are also shared with their own infants. Termination of breastfeeding and intrapartum antibiotic prophylaxis of mothers, which have the potential to affect microbial community composition, are associated with higher abundances of specific ARGs, the composition of which is largely shaped by bacterial phylogeny in the infant gut. Our results suggest that infants inherit the legacy of past antibiotic consumption of their mothers via transmission of genes, but microbiota composition still strongly impacts the overall resistance load.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- WHO World Health Organization. Antimicrobial resistance: global report on surveillance. Available at: http://www.who.int/drugresistance/documents/surveillancereport/en/. (2014). Accessed 27 Sept 2017.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

