Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis
- PMID: 30250259
- PMCID: PMC6155316
- DOI: 10.1038/s41467-018-06430-8
Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis
Abstract
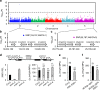
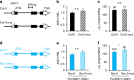
Soil water uptake by roots is a key component of plant performance and adaptation to adverse environments. Here, we use a genome-wide association analysis to identify the XYLEM NAC DOMAIN 1 (XND1) transcription factor as a negative regulator of Arabidopsis root hydraulic conductivity (Lpr). The distinct functionalities of a series of natural XND1 variants and a single nucleotide polymorphism that determines XND1 translation efficiency demonstrate the significance of XND1 natural variation at species-wide level. Phenotyping of xnd1 mutants and natural XND1 variants show that XND1 modulates Lpr through action on xylem formation and potential indirect effects on aquaporin function and that it diminishes drought stress tolerance. XND1 also mediates the inhibition of xylem formation by the bacterial elicitor flagellin and counteracts plant infection by the root pathogen Ralstonia solanacearum. Thus, genetic variation at XND1, and xylem differentiation contribute to resolving the major trade-off between abiotic and biotic stress resistance in Arabidopsis.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
The transcription factors DOF4.6 and XND1 jointly regulate root hydraulics and drought responses in Arabidopsis.Plant Cell. 2025 Apr 2;37(4):koaf083. doi: 10.1093/plcell/koaf083. Plant Cell. 2025. PMID: 40262770
-
Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis.Plant Cell. 2012 Sep;24(9):3823-37. doi: 10.1105/tpc.112.103374. Epub 2012 Sep 28. Plant Cell. 2012. PMID: 23023171 Free PMC article.
-
XYLEM NAC DOMAIN1, an angiosperm NAC transcription factor, inhibits xylem differentiation through conserved motifs that interact with RETINOBLASTOMA-RELATED.New Phytol. 2017 Oct;216(1):76-89. doi: 10.1111/nph.14704. Epub 2017 Jul 25. New Phytol. 2017. PMID: 28742236
-
XND1 Regulates Secondary Wall Deposition in Xylem Vessels through the Inhibition of VND Functions.Plant Cell Physiol. 2021 Mar 25;62(1):53-65. doi: 10.1093/pcp/pcaa140. Plant Cell Physiol. 2021. PMID: 33764471
-
Understanding the root xylem plasticity for designing resilient crops.Plant Cell Environ. 2022 Mar;45(3):664-676. doi: 10.1111/pce.14245. Epub 2021 Dec 31. Plant Cell Environ. 2022. PMID: 34971462 Free PMC article. Review.
Cited by
-
Comprehensive analysis of pathogen-responsive wheat NAC transcription factors: new candidates for crop improvement.G3 (Bethesda). 2022 Nov 4;12(11):jkac247. doi: 10.1093/g3journal/jkac247. G3 (Bethesda). 2022. PMID: 36130261 Free PMC article.
-
Root architecture and hydraulics converge for acclimation to changing water availability.Nat Plants. 2020 Jul;6(7):744-749. doi: 10.1038/s41477-020-0684-5. Epub 2020 Jun 29. Nat Plants. 2020. PMID: 32601421 Review.
-
Roots' Drought Adaptive Traits in Crop Improvement.Plants (Basel). 2022 Aug 30;11(17):2256. doi: 10.3390/plants11172256. Plants (Basel). 2022. PMID: 36079644 Free PMC article. Review.
-
Gibberellin Promotes Sweetpotato Root Vascular Lignification and Reduces Storage-Root Formation.Front Plant Sci. 2019 Nov 15;10:1320. doi: 10.3389/fpls.2019.01320. eCollection 2019. Front Plant Sci. 2019. PMID: 31849998 Free PMC article.
-
Current Understanding of the Genetics and Molecular Mechanisms Regulating Wood Formation in Plants.Genes (Basel). 2022 Jun 30;13(7):1181. doi: 10.3390/genes13071181. Genes (Basel). 2022. PMID: 35885964 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

