Escherichia coli Cells Exposed to Lethal Doses of Electron Beam Irradiation Retain Their Ability to Propagate Bacteriophages and Are Metabolically Active
- PMID: 30250460
- PMCID: PMC6139317
- DOI: 10.3389/fmicb.2018.02138
Escherichia coli Cells Exposed to Lethal Doses of Electron Beam Irradiation Retain Their Ability to Propagate Bacteriophages and Are Metabolically Active
Abstract
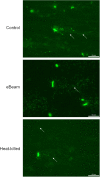
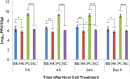
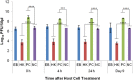
Reports in the literature suggest that bacteria exposed to lethal doses of ionizing radiation, i.e., electron beams, are unable to replicate yet they remain metabolically active. To investigate this phenomenon further, we electron beam irradiated Escherichia coli cells to a lethal dose and measured their membrane integrity, metabolic activity, ATP levels and overall cellular functionality via bacteriophage infection. We also visualized the DNA double-strand breaks in the cells. We used non-irradiated (live) and heat-killed cells as positive and negative controls, respectively. Our results show that the membrane integrity of E. coli cells is maintained and that the cells remain metabolically active up to 9 days post-irradiation when stored at 4°C. The ATP levels in lethally irradiated cells are similar to non-irradiated control cells. We also visualized extensive DNA damage within the cells and confirmed their cellular functionality based on their ability to propagate bacteriophages for up to 9 days post-irradiation. Overall, our findings indicate that lethally irradiated E. coli cells resemble live non-irradiated cells more closely than heat-killed (dead) cells.
Keywords: DNA damage; bacteria; bacteriophages; electron beam; ionizing radiation.
Figures


Similar articles
-
Radiation resistance and injury in starved Escherichia coli O157:H7 treated with electron-beam irradiation in 0.85% saline and in apple juice.Foodborne Pathog Dis. 2014 Nov;11(11):900-6. doi: 10.1089/fpd.2014.1782. Foodborne Pathog Dis. 2014. PMID: 25393670
-
Response of thyroid follicular cells to gamma irradiation compared to proton irradiation. I. Initial characterization of DNA damage, micronucleus formation, apoptosis, cell survival, and cell cycle phase redistribution.Radiat Res. 2001 Jan;155(1 Pt 1):32-42. doi: 10.1667/0033-7587(2001)155[0032:rotfct]2.0.co;2. Radiat Res. 2001. PMID: 11121213
-
Increased sensitivity to sparsely ionizing radiation due to excessive base excision in clustered DNA damage sites in Escherichia coli.Int J Radiat Biol. 2005 Feb;81(2):115-23. doi: 10.1080/09553000500103009. Int J Radiat Biol. 2005. PMID: 16019921
-
The relative contributions of DNA strand breaks, base damage and clustered lesions to the loss of DNA functionality induced by ionizing radiation.Radiat Res. 2014 Jan;181(1):99-110. doi: 10.1667/RR13450.1. Epub 2014 Jan 7. Radiat Res. 2014. PMID: 24397439
-
Systemic mechanisms and effects of ionizing radiation: A new 'old' paradigm of how the bystanders and distant can become the players.Semin Cancer Biol. 2016 Jun;37-38:77-95. doi: 10.1016/j.semcancer.2016.02.002. Epub 2016 Feb 9. Semin Cancer Biol. 2016. PMID: 26873647 Review.
Cited by
-
relA Inactivation Converts Sulfonamides Into Bactericidal Compounds.Front Microbiol. 2021 Sep 27;12:698468. doi: 10.3389/fmicb.2021.698468. eCollection 2021. Front Microbiol. 2021. PMID: 34646242 Free PMC article.
-
Imaging of Hydrated and Living Cells in Transmission Electron Microscope: Summary, Challenges, and Perspectives.ACS Nano. 2025 Apr 8;19(13):12710-12733. doi: 10.1021/acsnano.5c00871. Epub 2025 Mar 29. ACS Nano. 2025. PMID: 40156542 Free PMC article. Review.
-
Effect of Pefloxacin on Clostridioides difficile R20291 Persister Cells Formation.Antibiotics (Basel). 2025 Jun 20;14(7):628. doi: 10.3390/antibiotics14070628. Antibiotics (Basel). 2025. PMID: 40723930 Free PMC article.
-
A Comparative Analysis of the Metabolomic Response of Electron Beam Inactivated E. coli O26:H11 and Salmonella Typhimurium ATCC 13311.Front Microbiol. 2019 Apr 9;10:694. doi: 10.3389/fmicb.2019.00694. eCollection 2019. Front Microbiol. 2019. PMID: 31024484 Free PMC article.
-
Low-Energy Electron Irradiation Efficiently Inactivates the Gram-Negative Pathogen Rodentibacter pneumotropicus-A New Method for the Generation of Bacterial Vaccines with Increased Efficacy.Vaccines (Basel). 2020 Mar 2;8(1):113. doi: 10.3390/vaccines8010113. Vaccines (Basel). 2020. PMID: 32121656 Free PMC article.
References
-
- Ackermann H.-W. (2006). “Classification of bacteriophages,” in The Bacteriophages ed. Calendar R. (New York, NY: Oxford University Press; ) 8–16.
-
- Adams M. H. (1959). Bacteriophages. New York, NY: Interscience Publishers, Inc.
LinkOut - more resources
Full Text Sources
Other Literature Sources

