On the Evolution and Function of Plasmodium vivax Reticulocyte Binding Surface Antigen (pvrbsa)
- PMID: 30250483
- PMCID: PMC6139305
- DOI: 10.3389/fgene.2018.00372
On the Evolution and Function of Plasmodium vivax Reticulocyte Binding Surface Antigen (pvrbsa)
Abstract
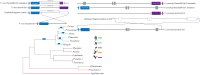
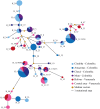
The RBSA protein is encoded by a gene described in Plasmodium species having tropism for reticulocytes. Since this protein is antigenic in natural infections and can bind to target cells, it has been proposed as a potential candidate for an anti-Plasmodium vivax vaccine. However, genetic diversity (a challenge which must be overcome for ensuring fully effective vaccine design) has not been described at this locus. Likewise, the minimum regions mediating specific parasite-host interaction have not been determined. This is why the rbsa gene's evolutionary history is being here described, as well as the P. vivax rbsa (pvrbsa) genetic diversity and the specific regions mediating parasite adhesion to reticulocytes. Unlike what has previously been reported, rbsa was also present in several parasite species belonging to the monkey-malaria clade; paralogs were also found in Plasmodium parasites invading reticulocytes. The pvrbsa locus had less diversity than other merozoite surface proteins where natural selection and recombination were the main evolutionary forces involved in causing the observed polymorphism. The N-terminal end (PvRBSA-A) was conserved and under functional constraint; consequently, it was expressed as recombinant protein for binding assays. This protein fragment bound to reticulocytes whilst the C-terminus, included in recombinant PvRBSA-B (which was not under functional constraint), did not. Interestingly, two PvRBSA-A-derived peptides were able to inhibit protein binding to reticulocytes. Specific conserved and functionally important peptides within PvRBSA-A could thus be considered when designing a fully-effective vaccine against P. vivax.
Keywords: Plasmodium vivax; antimalarial vaccine; evolutionary forces; genetic diversity; parasite–host interaction; protein biding; rbsa.
Figures


Similar articles
-
Characterising PvRBSA: an exclusive protein from Plasmodium species infecting reticulocytes.Parasit Vectors. 2017 May 18;10(1):243. doi: 10.1186/s13071-017-2185-6. Parasit Vectors. 2017. PMID: 28521840 Free PMC article.
-
Plasmodium vivax Merozoite Surface Protein 1 Paralog as a Mediator of Parasite Adherence to Reticulocytes.Infect Immun. 2018 Aug 22;86(9):e00239-18. doi: 10.1128/IAI.00239-18. Print 2018 Sep. Infect Immun. 2018. PMID: 29967091 Free PMC article.
-
Identifying Potential Plasmodium vivax Sporozoite Stage Vaccine Candidates: An Analysis of Genetic Diversity and Natural Selection.Front Genet. 2018 Jan 25;9:10. doi: 10.3389/fgene.2018.00010. eCollection 2018. Front Genet. 2018. PMID: 29422913 Free PMC article.
-
Plasmodium vivax in vitro continuous culture: the spoke in the wheel.Malar J. 2018 Aug 20;17(1):301. doi: 10.1186/s12936-018-2456-5. Malar J. 2018. PMID: 30126427 Free PMC article. Review.
-
Reticulocytes: Plasmodium vivax target cells.Biol Cell. 2013 Jun;105(6):251-60. doi: 10.1111/boc.201200093. Epub 2013 May 2. Biol Cell. 2013. PMID: 23458497 Review.
Cited by
-
The direct binding of Plasmodium vivax AMA1 to erythrocytes defines a RON2-independent invasion pathway.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2215003120. doi: 10.1073/pnas.2215003120. Epub 2022 Dec 28. Proc Natl Acad Sci U S A. 2023. PMID: 36577076 Free PMC article.
-
Plasmodium vivax: the potential obstacles it presents to malaria elimination and eradication.Trop Dis Travel Med Vaccines. 2022 Dec 15;8(1):27. doi: 10.1186/s40794-022-00185-3. Trop Dis Travel Med Vaccines. 2022. PMID: 36522671 Free PMC article. Review.
-
Whole genome sequencing of Plasmodium vivax isolates reveals frequent sequence and structural polymorphisms in erythrocyte binding genes.PLoS Negl Trop Dis. 2020 Oct 12;14(10):e0008234. doi: 10.1371/journal.pntd.0008234. eCollection 2020 Oct. PLoS Negl Trop Dis. 2020. PMID: 33044985 Free PMC article.
-
Evolution of Plasmodium vivax and resistance patterns for infection based on Duffy genotype and phenotype.Infez Med. 2023 Sep 1;31(3):350-358. doi: 10.53854/liim-3103-8. eCollection 2023. Infez Med. 2023. PMID: 37701383 Free PMC article. Review.
-
Babesia Bovis Ligand-Receptor Interaction: AMA-1 Contains Small Regions Governing Bovine Erythrocyte Binding.Int J Mol Sci. 2021 Jan 13;22(2):714. doi: 10.3390/ijms22020714. Int J Mol Sci. 2021. PMID: 33450807 Free PMC article.
References
-
- Arenas M., Posada D. (2014). “The influence of recombination on the estimation of selection from coding sequence alignments,” in Natural Selection: Methods and Applications, ed. Fares M. A. (Boca Raton, FL: CRC Press/Taylor & Francis; ), 112–125.
LinkOut - more resources
Full Text Sources
Other Literature Sources

