The functional diversity of Aurora kinases: a comprehensive review
- PMID: 30250494
- PMCID: PMC6146527
- DOI: 10.1186/s13008-018-0040-6
The functional diversity of Aurora kinases: a comprehensive review
Abstract
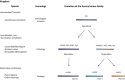
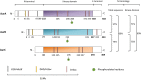
Aurora kinases are serine/threonine kinases essential for the onset and progression of mitosis. Aurora members share a similar protein structure and kinase activity, but exhibit distinct cellular and subcellular localization. AurA favors the G2/M transition by promoting centrosome maturation and mitotic spindle assembly. AurB and AurC are chromosome-passenger complex proteins, crucial for chromosome binding to kinetochores and segregation of chromosomes. Cellular distribution of AurB is ubiquitous, while AurC expression is mainly restricted to meiotically-active germ cells. In human tumors, all Aurora kinase members play oncogenic roles related to their mitotic activity and promote cancer cell survival and proliferation. Furthermore, AurA plays tumor-promoting roles unrelated to mitosis, including tumor stemness, epithelial-to-mesenchymal transition and invasion. In this review, we aim to understand the functional interplay of Aurora kinases in various types of human cells, including tumor cells. The understanding of the functional diversity of Aurora kinases could help to evaluate their relevance as potential therapeutic targets in cancer.
Keywords: Aurora kinase; Cancer; Mitosis.
Figures
References
-
- National Institute of General Medicine. Mitosis phases image. https://publications.nigms.nih.gov/insidethecell/ch4_phases_allbig.html. 2018. p. 1. https://publications.nigms.nih.gov/order/pubs_gateway.html. Accessed 11 Jun 2018.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

