A Pharmacochaperone-Based High-Throughput Screening Assay for the Discovery of Chemical Probes of Orphan Receptors
- PMID: 30251873
- PMCID: PMC6207161
- DOI: 10.1089/adt.2018.868
A Pharmacochaperone-Based High-Throughput Screening Assay for the Discovery of Chemical Probes of Orphan Receptors
Abstract
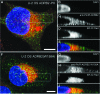
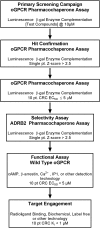
G-protein-coupled receptors (GPCRs) have varying and diverse physiological roles, transmitting signals from a range of stimuli, including light, chemicals, peptides, and mechanical forces. More than 130 GPCRs are orphan receptors (i.e., their endogenous ligands are unknown), representing a large untapped reservoir of potential therapeutic targets for pharmaceutical intervention in a variety of diseases. Current deorphanization approaches are slow, laborious, and usually require some in-depth knowledge about the receptor pharmacology. In this study we describe a cell-based assay to identify small molecule probes of orphan receptors that requires no a priori knowledge of receptor pharmacology. Built upon the concept of pharmacochaperones, where cell-permeable small molecules facilitate the trafficking of mutant receptors to the plasma membrane, the simple and robust technology is readily accessible by most laboratories and is amenable to high-throughput screening. The assay consists of a target harboring a synthetic point mutation that causes retention of the target in the endoplasmic reticulum. Coupled with a beta-galactosidase enzyme-fragment complementation reporter system, the assay identifies compounds that act as pharmacochaperones causing forward trafficking of the mutant GPCR. The assay can identify compounds with varying mechanisms of action including agonists and antagonists. A universal positive control compound circumvents the need for a target-specific ligand. The veracity of the approach is demonstrated using the beta-2-adrenergic receptor. Together with other existing assay technologies to validate the signaling pathways and the specificity of ligands identified, this pharmacochaperone-based approach can accelerate the identification of ligands for these potentially therapeutically useful receptors.
Keywords: chemical biology; deorphanization; orphan GPCR; pharmacochaperone; proteasome inhibitor.
Conflict of interest statement
J.L. and D.B. are employees of Eurofins DiscoverX. At the time this work was initiated, T.W. was employed by Eurofins DiscoverX. He is currently employed by PrimityBio.
Figures





Similar articles
-
Screening β-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors.J Biomol Screen. 2013 Jun;18(5):599-609. doi: 10.1177/1087057113475480. Epub 2013 Feb 8. J Biomol Screen. 2013. PMID: 23396314
-
Discovery of selective probes and antagonists for G-protein-coupled receptors FPR/FPRL1 and GPR30.Curr Top Med Chem. 2009;9(13):1227-36. doi: 10.2174/156802609789753608. Curr Top Med Chem. 2009. PMID: 19807662 Free PMC article. Review.
-
GPCR profiling: from hits to leads and from genotype to phenotype.Drug Discov Today Technol. 2015 Nov;18:30-7. doi: 10.1016/j.ddtec.2015.10.005. Epub 2015 Nov 6. Drug Discov Today Technol. 2015. PMID: 26723890 Review.
-
Identification of signal transduction pathways used by orphan g protein-coupled receptors.Assay Drug Dev Technol. 2003 Apr;1(2):239-49. doi: 10.1089/15406580360545053. Assay Drug Dev Technol. 2003. PMID: 15090189
-
Pharmacology of Free Fatty Acid Receptors and Their Allosteric Modulators.Int J Mol Sci. 2021 Feb 10;22(4):1763. doi: 10.3390/ijms22041763. Int J Mol Sci. 2021. PMID: 33578942 Free PMC article. Review.
Cited by
-
Soluble CD13 induces inflammatory arthritis by activating the bradykinin receptor B1.J Clin Invest. 2022 Jun 1;132(11):e151827. doi: 10.1172/JCI151827. J Clin Invest. 2022. PMID: 35439173 Free PMC article.
-
Orphan G protein-coupled receptors: the ongoing search for a home.Front Pharmacol. 2024 Feb 29;15:1349097. doi: 10.3389/fphar.2024.1349097. eCollection 2024. Front Pharmacol. 2024. PMID: 38495099 Free PMC article. Review.
References
-
- Hopkins AL, Groom CR: The druggable genome. Nat Rev Drug Discov 2002;1:727–730 - PubMed
-
- Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schioth HB: Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics 2006;88:263–273 - PubMed
-
- Fredriksson R, Schioth HB: The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol 2005;67:1414–1425 - PubMed
-
- Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, Yu FH: Too many roads not taken. Nature 2011;470:163–165 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

