A Chemical Signature for Cytidine Acetylation in RNA
- PMID: 30252461
- PMCID: PMC8054311
- DOI: 10.1021/jacs.8b06636
A Chemical Signature for Cytidine Acetylation in RNA
Abstract
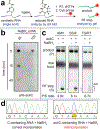
N4-acetylcytidine (ac4C) is a highly conserved modified RNA nucleobase whose formation is catalyzed by the disease-associated N-acetyltransferase 10 (NAT10). Here we report a sensitive chemical method to localize ac4C in RNA. Specifically, we characterize the susceptibility of ac4C to borohydride-based reduction and show this reaction can cause introduction of noncognate base pairs during reverse transcription (RT). Combining borohydride-dependent misincorporation with ac4C's known base-sensitivity provides a unique chemical signature for this modified nucleobase. We show this unique reactivity can be used to quantitatively analyze cellular RNA acetylation, study adapters responsible for ac4C targeting, and probe the timing of RNA acetylation during ribosome biogenesis. Overall, our studies provide a chemical foundation for defining an expanding landscape of cytidine acetyltransferase activity and its impact on biology and disease.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

