Moonlighting proteins: putting the spotlight on enzymes
- PMID: 30252596
- PMCID: PMC6204816
- DOI: 10.1080/15592324.2018.1517075
Moonlighting proteins: putting the spotlight on enzymes
Abstract
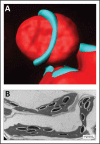
AROGENATE DEHAYDRATASE2 (ADT2) is a member of the Arabidopsis thaliana ADT family. All members of this family act as arogenate dehydratases in phenylalanine biosynthesis, decarboxylating/dehydrating arogenate to phenylalanine. ADT2 is detected in stromules, and as a ring around the equatorial plane of dividing chloroplasts, indicating it has a second, non-enzymatic function in chloroplast division. Here, we provide further evidence for this alternative role of ADT2. First, we demonstrate that ADT2 and FtsZ co-localize around the equatorial plane at the same time. Second, FtsZ expression in an adt2 mutant was analyzed, as well as ADT2 expression in three Arabidopsis chloroplast division mutants, ACCUMULATION AND REPLICATION OF CHLOROPLASTS3 (ARC3), ARC5 and ARC6. In arc3 and arc6 mutants, ADT2 is misexpressed and resembles the expression of FtsZ in the same mutants. However, in the arc5 mutant, ADT2 ring positioning is observed at constriction points indicating proper relative timing. ADT2 expression in the arc mutants shows that the role of ADT2 in chloroplast division occurs prior to ARC5, but is dependent on ARC3 and ARC6. Abbreviations used: ADT: arogenate dehydratase, ARC: accumulation and replication of chloroplasts, CFP: cyan fluorescent protein, dpi: days post infiltration, FtsZ: filamentous temperature sensitive Z, PD: plastid division, Phe: phenylalanine, YFP: yellow fluorescent protein.
Keywords: Arc mutants; FtsZ; arogenate dehydratase; chloroplast division; gene families; moonlighting proteins; phenylalanine biosynthesis.
Figures

References
-
- Laohavisit A, Davies JM. Multifunctional annexins. Plant Science. 2009;177:532. doi: 10.1016/j.plantsci.2009.09.008. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
