Molecular analysis of the chromosomal 16S rRNA gene and vapA plasmid gene of Polish field strains of R. equi
- PMID: 30252885
- PMCID: PMC6155501
- DOI: 10.1371/journal.pone.0204024
Molecular analysis of the chromosomal 16S rRNA gene and vapA plasmid gene of Polish field strains of R. equi
Abstract
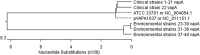
Rhodococcus equi (R. hoagii) is an opportunistic pathogen commonly found in foals up to 6 months old and animal environment. The R. equi genome contains genetically stable chromosomal DNA and an 80-90 kb plasmid containing vapA gene, responsible for virulence. Most reports from around the world focus on the determination of R. equi plasmid profiles. Few studies have attempted to determine differences in nucleotide sequences between virulent strains of R. equi isolated from foals and breeding environment. The aim of the study was to perform a molecular analysis of a fragment of the chromosomal gene encoding the 16S rRNA subunit and the vapA plasmid gene of virulent R. equi strains isolated on Polish studs from foals and from the breeding environment of horses. The sequencing method was used to compare the primary structure of fragments of the chromosomal and plasmid DNA of the virulent R. equi strains. The sequences of 22 clinical and 18 environmental R. equi isolates were compared with the sequences of the gene fragments of reference strains available in the NCBI GenBank database. All sequenced 16S rRNA amplicons of Polish field strains were identical and showed 99.5% similarity to the four randomly selected sequences of this gene fragment in the GenBank database. The results confirm that fragments of the 16S rRNA gene of R. equi strains are highly conserved and do not undergo variation in field conditions. Analysis of the sequencing results for the vapA gene fragment of the strains used in our study revealed two polymorphic variants and clear differences between the sequences of strains isolated from foals and from soil samples. Presumably, R. equi strains present in the breeding environment are more exposed than clinical strains to adverse external factors. This may result in changes in the DNA sequence due to natural selection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Plasmid Profiles of Virulent Rhodococcus equi Strains Isolated from Infected Foals in Poland.PLoS One. 2016 Apr 13;11(4):e0152887. doi: 10.1371/journal.pone.0152887. eCollection 2016. PLoS One. 2016. PMID: 27074033 Free PMC article.
-
Prevalence of virulent Rhodococcus equi in soil from five R. equi-endemic horse-breeding farms and restriction fragment length polymorphisms of virulence plasmids in isolates from soil and infected foals in Texas.J Vet Diagn Invest. 2001 Nov;13(6):489-94. doi: 10.1177/104063870101300606. J Vet Diagn Invest. 2001. PMID: 11724139
-
Characterization of virulence plasmid types in Rhodococcus equi isolates from foals, pigs, humans and soil in Hungary.Vet Microbiol. 2002 Sep 24;88(4):377-84. doi: 10.1016/s0378-1135(02)00157-8. Vet Microbiol. 2002. PMID: 12220812
-
Short review: Geographical distribution of equine-associated pVAPA plasmids in Rhodococcus equi in the world.Vet Microbiol. 2023 Dec;287:109919. doi: 10.1016/j.vetmic.2023.109919. Epub 2023 Nov 21. Vet Microbiol. 2023. PMID: 38000208 Review.
-
Rhodococcus equi: the many facets of a pathogenic actinomycete.Vet Microbiol. 2013 Nov 29;167(1-2):9-33. doi: 10.1016/j.vetmic.2013.06.016. Epub 2013 Jul 5. Vet Microbiol. 2013. PMID: 23993705 Review.
Cited by
-
Inoculation With a Microbe Isolated From the Negev Desert Enhances Corn Growth.Front Microbiol. 2020 Jun 19;11:1149. doi: 10.3389/fmicb.2020.01149. eCollection 2020. Front Microbiol. 2020. PMID: 32636811 Free PMC article.
References
-
- Kämpfer P, Dott W, Martin K, Glaeser SP. Rhodococcus defluvii sp. nov., isolated from wastewater of a bioreactor and formal proposal to reclassify [Corynebacterium hoagii] and Rhodococcus equi as Rhodococcus hoagii comb. nov. Int J Syst Evol Microbiol. 2014;64: 755–761. 10.1099/ijs.0.053322-0 - DOI - PubMed
-
- Duquesne F, Hébert L, Sévin C, Breuil MF, Tapprest J, Laugier C, et al. Analysis of plasmid diversity in 96 Rhodococcus equi strains isolated in Normandy (France) and sequencing of the 87-kb type I virulence plasmid. FEMS Microbiol Lett. 2010;311: 76–81. 10.1111/j.1574-6968.2010.02070.x - DOI - PubMed
-
- Muscatello G, Leadon DP, Klayt M, Ocampo-Sosa A, Lewis DA, Fogarty U, et al. Rhodococcus equi infection in foals: the science of 'rattles'. Equine Vet J. 2007;39: 470–478. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

