Using correlated motions to determine sufficient sampling times for molecular dynamics
- PMID: 30253618
- PMCID: PMC6325644
- DOI: 10.1103/PhysRevE.98.023307
Using correlated motions to determine sufficient sampling times for molecular dynamics
Abstract
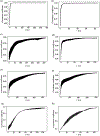
Here we present a time-dependent correlation method that provides insight into how long a system takes to grow into its equal-time (Pearson) correlation. We also show a usage of an extant time-lagged correlation method that indicates the time for parts of a system to become decorrelated, relative to equal-time correlation. Given a completed simulation (or set of simulations), these tools estimate (i) how long of a simulation of the same system would be sufficient to observe the same correlated motions, (ii) if patterns of observed correlated motions indicate events beyond the timescale of the simulation, and (iii) how long of a simulation is needed to observe these longer timescale events. We view this method as a decision-support tool that will aid researchers in determining necessary sampling times. In principle, this tool is extendable to any multidimensional time series data with a notion of correlated fluctuations; however, here we limit our discussion to data from molecular-dynamics simulations.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
