Comparison of RNA-seq and microarray platforms for splice event detection using a cross-platform algorithm
- PMID: 30253752
- PMCID: PMC6156849
- DOI: 10.1186/s12864-018-5082-2
Comparison of RNA-seq and microarray platforms for splice event detection using a cross-platform algorithm
Abstract
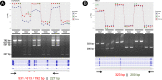
Background: RNA-seq is a reference technology for determining alternative splicing at genome-wide level. Exon arrays remain widely used for the analysis of gene expression, but show poor validation rate with regard to splicing events. Commercial arrays that include probes within exon junctions have been developed in order to overcome this problem. We compare the performance of RNA-seq (Illumina HiSeq) and junction arrays (Affymetrix Human Transcriptome array) for the analysis of transcript splicing events. Three different breast cancer cell lines were treated with CX-4945, a drug that severely affects splicing. To enable a direct comparison of the two platforms, we adapted EventPointer, an algorithm that detects and labels alternative splicing events using junction arrays, to work also on RNA-seq data. Common results and discrepancies between the technologies were validated and/or resolved by over 200 PCR experiments.
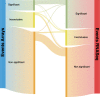
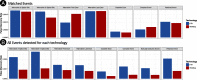
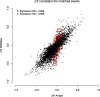
Results: As might be expected, RNA-seq appears superior in cases where the technologies disagree and is able to discover novel splicing events beyond the limitations of physical probe-sets. We observe a high degree of coherence between the two technologies, however, with correlation of EventPointer results over 0.90. Through decimation, the detection power of the junction arrays is equivalent to RNA-seq with up to 60 million reads.
Conclusions: Our results suggest, therefore, that exon-junction arrays are a viable alternative to RNA-seq for detection of alternative splicing events when focusing on well-described transcriptional regions.
Keywords: Alternative splicing; Microarrays; RNA-seq.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable as cell lines were obtained from commercially available suppliers. Cell lines MDA-MB-231 and MDA-MB-468 were obtained from ATCC (Manassas, VA) with identification numbers HTB-26 and HT-132 respectively and cell line SUM149 was purchased from Asterand plc (Detroit, MI).
Consent for publication
Not applicable.
Competing interests
A. Rubio, J.P. Romero and F. Carazo are being funded by Affymetrix in an independent project. The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
EventPointer: an effective identification of alternative splicing events using junction arrays.BMC Genomics. 2016 Jun 17;17:467. doi: 10.1186/s12864-016-2816-x. BMC Genomics. 2016. PMID: 27315794 Free PMC article.
-
RNA sequencing and transcriptome arrays analyses show opposing results for alternative splicing in patient derived samples.BMC Genomics. 2017 Jun 6;18(1):443. doi: 10.1186/s12864-017-3819-y. BMC Genomics. 2017. PMID: 28587590 Free PMC article.
-
Read-Split-Run: an improved bioinformatics pipeline for identification of genome-wide non-canonical spliced regions using RNA-Seq data.BMC Genomics. 2016 Aug 22;17 Suppl 7(Suppl 7):503. doi: 10.1186/s12864-016-2896-7. BMC Genomics. 2016. PMID: 27556805 Free PMC article.
-
Alternative splicing, RNA-seq and drug discovery.Drug Discov Today. 2019 Jun;24(6):1258-1267. doi: 10.1016/j.drudis.2019.03.030. Epub 2019 Apr 4. Drug Discov Today. 2019. PMID: 30953866 Review.
-
Genomic analysis of RNA alternative splicing in cancers.Front Biosci. 2007 May 1;12:4034-41. doi: 10.2741/2369. Front Biosci. 2007. PMID: 17485356 Review.
Cited by
-
Differential Expression Analyses on Human Aortic Tissue Reveal Novel Genes and Pathways Associated With Abdominal Aortic Aneurysm Onset and Progression.J Am Heart Assoc. 2024 Dec 17;13(24):e036082. doi: 10.1161/JAHA.124.036082. Epub 2024 Dec 10. J Am Heart Assoc. 2024. PMID: 39655704 Free PMC article.
-
Circular RNAs and RNA Splice Variants as Biomarkers for Prognosis and Therapeutic Response in the Liquid Biopsies of Lung Cancer Patients.Front Genet. 2019 May 7;10:390. doi: 10.3389/fgene.2019.00390. eCollection 2019. Front Genet. 2019. PMID: 31134126 Free PMC article. Review.
-
Integration of CLIP experiments of RNA-binding proteins: a novel approach to predict context-dependent splicing factors from transcriptomic data.BMC Genomics. 2019 Jun 25;20(1):521. doi: 10.1186/s12864-019-5900-1. BMC Genomics. 2019. PMID: 31238884 Free PMC article.
-
Systematic Identification of Housekeeping Genes Possibly Used as References in Caenorhabditis elegans by Large-Scale Data Integration.Cells. 2020 Mar 24;9(3):786. doi: 10.3390/cells9030786. Cells. 2020. PMID: 32213971 Free PMC article.
-
The Study of Alternative Splicing Events in Human Induced Pluripotent Stem Cells From a Down's Syndrome Patient.Front Cell Dev Biol. 2021 Sep 30;9:661381. doi: 10.3389/fcell.2021.661381. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34660567 Free PMC article.
References
-
- Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, Davies C, Williams A, Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

