CdrA Interactions within the Pseudomonas aeruginosa Biofilm Matrix Safeguard It from Proteolysis and Promote Cellular Packing
- PMID: 30254118
- PMCID: PMC6156197
- DOI: 10.1128/mBio.01376-18
CdrA Interactions within the Pseudomonas aeruginosa Biofilm Matrix Safeguard It from Proteolysis and Promote Cellular Packing
Abstract
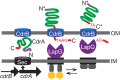
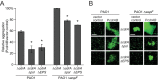
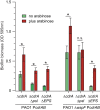
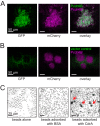
Biofilms are robust multicellular aggregates of bacteria that are encased in an extracellular matrix. Different bacterial species have been shown to use a range of biopolymers to build their matrices. Pseudomonas aeruginosa is a model organism for the laboratory study of biofilms, and past work has suggested that exopolysaccharides are a required matrix component. However, we found that expression of the matrix protein CdrA, in the absence of biofilm exopolysaccharides, allowed biofilm formation through the production of a CdrA-rich proteinaceous matrix. This represents a novel function for CdrA. Similar observations have been made for other species such as Escherichia coli and Staphylococcus aureus, which can utilize protein-dominant biofilm matrices. However, we found that these CdrA-containing matrices were susceptible to both exogenous and self-produced proteases. We previously reported that CdrA directly binds the biofilm matrix exopolysaccharide Psl. Now we have found that when CdrA bound to Psl, it was protected from proteolysis. Together, these results support the idea of the importance of multibiomolecular components in matrix stability and led us to propose a model in which CdrA-CdrA interactions can enhance cell-cell packing in an aggregate that is resistant to physical shear, while Psl-CdrA interactions enhance aggregate integrity in the presence of self-produced and exogenous proteases.IMPORTANCEPseudomonas aeruginosa forms multicellular aggregates or biofilms using both exopolysaccharides and the CdrA matrix adhesin. We showed for the first time that P. aeruginosa can use CdrA to build biofilms that do not require known matrix exopolysaccharides. It is appreciated that biofilm growth is protective against environmental assaults. However, little is known about how the interactions between individual matrix components aid in this protection. We found that interactions between CdrA and the exopolysaccharide Psl fortify the matrix by preventing CdrA proteolysis. When both components-CdrA and Psl-are part of the matrix, robust aggregates form that are tightly packed and protease resistant. These findings provide insight into how biofilms persist in protease-rich host environments.
Keywords: CdrA; Pseudomonas aeruginosa; Psl; biofilm; elastase; exopolysaccharides.
Copyright © 2018 Reichhardt et al.
Figures
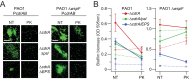

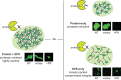
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
