RARα supports the development of Langerhans cells and langerin-expressing conventional dendritic cells
- PMID: 30254197
- PMCID: PMC6156335
- DOI: 10.1038/s41467-018-06341-8
RARα supports the development of Langerhans cells and langerin-expressing conventional dendritic cells
Abstract
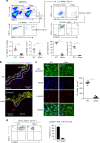
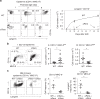
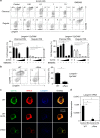
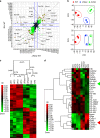
Langerhans cells (LC) are the prototype langerin-expressing dendritic cells (DC) that reside specifically in the epidermis, but langerin-expressing conventional DCs also reside in the dermis and other tissues, yet the factors that regulate their development are unclear. Because retinoic acid receptor alpha (RARα) is highly expressed by LCs, we investigate the functions of RARα and retinoic acid (RA) in regulating the langerin-expressing DCs. Here we show that the development of LCs from embryonic and bone marrow-derived progenitors and langerin+ conventional DCs is profoundly regulated by the RARα-RA axis. During LC differentiation, RARα is required for the expression of a LC-promoting transcription factor Runx3, but suppresses that of LC-inhibiting C/EBPβ. RARα promotes the development of LCs and langerin+ conventional DCs only in hypo-RA conditions, a function effectively suppressed at systemic RA levels. Our findings identify positive and negative regulatory mechanisms to tightly regulate the development of the specialized DC populations.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The dermis contains langerin+ dendritic cells that develop and function independently of epidermal Langerhans cells.J Exp Med. 2007 Dec 24;204(13):3119-31. doi: 10.1084/jem.20071724. Epub 2007 Dec 17. J Exp Med. 2007. PMID: 18086861 Free PMC article.
-
Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells.J Leukoc Biol. 2015 Apr;97(4):627-34. doi: 10.1189/jlb.1HI0714-351R. Epub 2014 Dec 16. J Leukoc Biol. 2015. PMID: 25516751 Free PMC article.
-
Langerhans Cell-Dendritic Cell Cross-Talk via Langerin and Hyaluronic Acid Mediates Antigen Transfer and Cross-Presentation of HIV-1.J Immunol. 2015 Aug 15;195(4):1763-73. doi: 10.4049/jimmunol.1402356. Epub 2015 Jul 13. J Immunol. 2015. PMID: 26170391
-
Langerin/CD207 sheds light on formation of birbeck granules and their possible function in Langerhans cells.Immunol Res. 2003;28(2):93-107. doi: 10.1385/IR:28:2:93. Immunol Res. 2003. PMID: 14610287 Review.
-
Langerhans cells and more: langerin-expressing dendritic cell subsets in the skin.Immunol Rev. 2010 Mar;234(1):120-41. doi: 10.1111/j.0105-2896.2009.00886.x. Immunol Rev. 2010. PMID: 20193016 Free PMC article. Review.
Cited by
-
MSC-like cells increase ability of monocyte-derived dendritic cells to polarize IL-17-/IL-10-producing T cells via CTLA-4.iScience. 2021 Mar 15;24(4):102312. doi: 10.1016/j.isci.2021.102312. eCollection 2021 Apr 23. iScience. 2021. PMID: 33855282 Free PMC article.
-
A ligand-independent fast function of RARα promotes exit from metabolic quiescence upon T cell activation and controls T cell differentiation.Mucosal Immunol. 2021 Jan;14(1):100-112. doi: 10.1038/s41385-020-0311-9. Epub 2020 Jun 9. Mucosal Immunol. 2021. PMID: 32518366 Free PMC article.
-
Skin Development and Disease: A Molecular Perspective.Curr Issues Mol Biol. 2024 Jul 30;46(8):8239-8267. doi: 10.3390/cimb46080487. Curr Issues Mol Biol. 2024. PMID: 39194704 Free PMC article. Review.
-
Effects of Anionic Liposome Delivery of All-Trans-Retinoic Acid on Neuroblastoma Cell Differentiation.Biomimetics (Basel). 2024 Apr 24;9(5):257. doi: 10.3390/biomimetics9050257. Biomimetics (Basel). 2024. PMID: 38786467 Free PMC article.
-
A role for whey acidic protein four-disulfide-core 12 (WFDC12) in the pathogenesis and development of psoriasis disease.Front Immunol. 2022 Sep 6;13:873720. doi: 10.3389/fimmu.2022.873720. eCollection 2022. Front Immunol. 2022. PMID: 36148224 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01AI080769/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)/International
- R01 AI080769/AI/NIAID NIH HHS/United States
- R01DK076616/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)/International
- R01 AI121302/AI/NIAID NIH HHS/United States
- R01 DK076616/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

