Deficiency of MecA in Streptococcus mutans Causes Major Defects in Cell Envelope Biogenesis, Cell Division, and Biofilm Formation
- PMID: 30254619
- PMCID: PMC6141683
- DOI: 10.3389/fmicb.2018.02130
Deficiency of MecA in Streptococcus mutans Causes Major Defects in Cell Envelope Biogenesis, Cell Division, and Biofilm Formation
Abstract
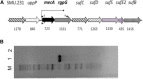
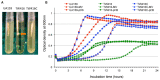
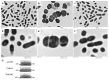
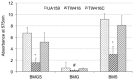
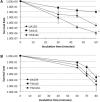
MecA is an adaptor protein that guides the ClpC/P-mediated proteolysis. A S. mutans MecA-deficient mutant was constructed by double-crossover allelic exchange and analyzed for the effects of such a deficiency on cell biology and biofilm formation. Unlike the wild-type, UA159, the mecA mutant, TW416, formed mucoid and smooth colonies, severely clumped in broth and had a reduced growth rate. Transmission electron microscopy analysis revealed that TW416 grows primarily in chains of giant "swollen" cells with multiple asymmetric septa, unlike the coccoid form of UA159. As compared to UA159, biofilm formation by TW416 was significantly reduced regardless of the carbohydrate sources used for growth (P < 0.001). Western blot analysis of TW416 whole cell lysates showed a reduced expression of the glucosyltransferase GtfC and GtfB, as well as the P1 and WapA adhesins providing an explanation for the defective biofilm formation of TW416. When analyzed by a colorimetric assay, the cell wall phosphate of the mutant murein sacculi was almost 20-fold lower than the parent strain (P < 0.001). Interestingly, however, when analyzed using immunoblotting of the murein sacculi preps with UA159 whole cell antiserum as a probe, TW416 was shown to possess significantly higher signal intensity as compared to the wild-type. There is also evidence that MecA in S. mutans is more than an adaptor protein, although how it modulates the bacterial pathophysiology, including cell envelope biogenesis, cell division, and biofilm formation awaits further investigation.
Keywords: Clp protease complex; MecA; Streptococcus mutans; biofilms; cell envelope biogenesis; stress tolerance response.
Figures




Similar articles
-
Impacts of a DUF2207 Family Protein on Streptococcus mutans Stress Tolerance Responses and Biofilm Formation.Microorganisms. 2023 Aug 1;11(8):1982. doi: 10.3390/microorganisms11081982. Microorganisms. 2023. PMID: 37630542 Free PMC article.
-
Deficiency of RgpG Causes Major Defects in Cell Division and Biofilm Formation, and Deficiency of LytR-CpsA-Psr Family Proteins Leads to Accumulation of Cell Wall Antigens in Culture Medium by Streptococcus mutans.Appl Environ Microbiol. 2017 Aug 17;83(17):e00928-17. doi: 10.1128/AEM.00928-17. Print 2017 Sep 1. Appl Environ Microbiol. 2017. PMID: 28687645 Free PMC article.
-
MecA in Streptococcus mutans is a multi-functional protein.mSphere. 2024 Dec 19;9(12):e0030824. doi: 10.1128/msphere.00308-24. Epub 2024 Nov 12. mSphere. 2024. PMID: 39530674 Free PMC article.
-
PBP1a-deficiency causes major defects in cell division, growth and biofilm formation by Streptococcus mutans.PLoS One. 2015 Apr 16;10(4):e0124319. doi: 10.1371/journal.pone.0124319. eCollection 2015. PLoS One. 2015. PMID: 25880908 Free PMC article.
-
Deficiency of PdxR in Streptococcus mutans affects vitamin B6 metabolism, acid tolerance response and biofilm formation.Mol Oral Microbiol. 2015 Aug;30(4):255-68. doi: 10.1111/omi.12090. Epub 2015 Jan 21. Mol Oral Microbiol. 2015. PMID: 25421565 Free PMC article.
Cited by
-
MecA: A Multifunctional ClpP-Dependent and Independent Regulator in Gram-Positive Bacteria.Mol Microbiol. 2025 May;123(5):433-438. doi: 10.1111/mmi.15356. Epub 2025 Mar 11. Mol Microbiol. 2025. PMID: 40070161 Review.
-
Streptococcus mutans requires mature rhamnose-glucose polysaccharides for proper pathophysiology, morphogenesis and cellular division.Mol Microbiol. 2019 Sep;112(3):944-959. doi: 10.1111/mmi.14330. Epub 2019 Jul 12. Mol Microbiol. 2019. PMID: 31210392 Free PMC article.
-
Prevalence and Impact of Biofilms on Bloodstream and Urinary Tract Infections: A Systematic Review and Meta-Analysis.Antibiotics (Basel). 2021 Jul 8;10(7):825. doi: 10.3390/antibiotics10070825. Antibiotics (Basel). 2021. PMID: 34356749 Free PMC article.
-
Impacts of a DUF2207 Family Protein on Streptococcus mutans Stress Tolerance Responses and Biofilm Formation.Microorganisms. 2023 Aug 1;11(8):1982. doi: 10.3390/microorganisms11081982. Microorganisms. 2023. PMID: 37630542 Free PMC article.
-
Antimicrobial Activity of Cinnamaldehyde on Streptococcus mutans Biofilms.Front Microbiol. 2019 Sep 25;10:2241. doi: 10.3389/fmicb.2019.02241. eCollection 2019. Front Microbiol. 2019. PMID: 31608045 Free PMC article.
References
-
- Aspiras M. B., Ellen R. P., Cvitkovitch D. G. (2004). ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol. Lett. 238 167–174. - PubMed
-
- Besingi R. N., Wenderska I. B., Senadheera D. B., Cvitkovitch D. G., Long J. R., Wen Z. T., et al. (2017). Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c. Microbiology 163 488–501. 10.1099/mic.0.000443 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

