Adenoviromics: Mining the Human Adenovirus Species D Genome
- PMID: 30254627
- PMCID: PMC6141750
- DOI: 10.3389/fmicb.2018.02178
Adenoviromics: Mining the Human Adenovirus Species D Genome
Erratum in
-
Corrigendum: Adenoviromics: Mining the Human Adenovirus Species D Genome.Front Microbiol. 2018 Dec 4;9:3005. doi: 10.3389/fmicb.2018.03005. eCollection 2018. Front Microbiol. 2018. PMID: 30555456 Free PMC article.
Abstract
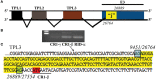
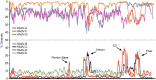
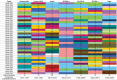
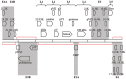
Human adenovirus (HAdV) infections cause disease world-wide. Whole genome sequencing has now distinguished 90 distinct genotypes in 7 species (A-G). Over half of these 90 HAdVs fall within species D, with essentially all of the HAdV-D whole genome sequences generated in the last decade. Herein, we describe recent new findings made possible by mining of this expanded genome database, and propose future directions to elucidate new functional elements and new functions for previously known viral components.
Keywords: adenovirus; evolution; genome; interactome; transcription factor.
Figures


References
-
- Andersson M., McMichael A., Peterson P. A. (1987). Reduced allorecognition of adenovirus-2 infected cells. J. Immunol. 138, 3960–3966. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

