Compressive force induces reversible chromatin condensation and cell geometry-dependent transcriptional response
- PMID: 30256731
- PMCID: PMC6333178
- DOI: 10.1091/mbc.E18-04-0256
Compressive force induces reversible chromatin condensation and cell geometry-dependent transcriptional response
Abstract
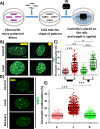
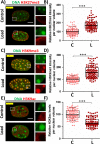
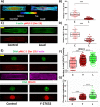
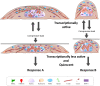
Fibroblasts exhibit heterogeneous cell geometries in tissues and integrate both mechanical and biochemical signals in their local microenvironment to regulate genomic programs via chromatin remodelling. While in connective tissues fibroblasts experience tensile and compressive forces (CFs), the role of compressive forces in regulating cell behavior and, in particular, the impact of cell geometry in modulating transcriptional response to such extrinsic mechanical forces is unclear. Here we show that CF on geometrically well-defined mouse fibroblast cells reduces actomyosin contractility and shuttles histone deacetylase 3 (HDAC3) into the nucleus. HDAC3 then triggers an increase in the heterochromatin content by initiating removal of acetylation marks on the histone tails. This suggests that, in response to CF, fibroblasts condense their chromatin and enter into a transcriptionally less active and quiescent states as also revealed by transcriptome analysis. On removal of CF, the alteration in chromatin condensation was reversed. We also present a quantitative model linking CF-dependent changes in actomyosin contractility leading to chromatin condensation. Further, transcriptome analysis also revealed that the transcriptional response of cells to CF was geometry dependent. Collectively, our results suggest that CFs induce chromatin condensation and geometry-dependent differential transcriptional response in fibroblasts that allows maintenance of tissue homeostasis.
Figures




References
-
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, et al (2006). Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res , 707–733. - PubMed
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data, version 0.11.4. Available online at www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed April 2017).
-
- Anwar T, Sen B, Aggarwal S, Nath R, Pathak N, Katoch A, Aiyaz M, Trehanpati N, Khosla S, Ramakrishna G. (2018). Differentially regulated gene expression in quiescence versus senescence and identification of ARID5A as a quiescence associated marker. J Cell Physiol , 3695–3712. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

