Genetic basis of thermal plasticity variation in Drosophila melanogaster body size
- PMID: 30256798
- PMCID: PMC6175520
- DOI: 10.1371/journal.pgen.1007686
Genetic basis of thermal plasticity variation in Drosophila melanogaster body size
Abstract
Body size is a quantitative trait that is closely associated to fitness and under the control of both genetic and environmental factors. While developmental plasticity for this and other traits is heritable and under selection, little is known about the genetic basis for variation in plasticity that can provide the raw material for its evolution. We quantified genetic variation for body size plasticity in Drosophila melanogaster by measuring thorax and abdomen length of females reared at two temperatures from a panel representing naturally segregating alleles, the Drosophila Genetic Reference Panel (DGRP). We found variation between genotypes for the levels and direction of thermal plasticity in size of both body parts. We then used a Genome-Wide Association Study (GWAS) approach to unravel the genetic basis of inter-genotype variation in body size plasticity, and used different approaches to validate selected QTLs and to explore potential pleiotropic effects. We found mostly "private QTLs", with little overlap between the candidate loci underlying variation in plasticity for thorax versus abdomen size, for different properties of the plastic response, and for size versus size plasticity. We also found that the putative functions of plasticity QTLs were diverse and that alleles for higher plasticity were found at lower frequencies in the target population. Importantly, a number of our plasticity QTLs have been targets of selection in other populations. Our data sheds light onto the genetic basis of inter-genotype variation in size plasticity that is necessary for its evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

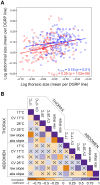

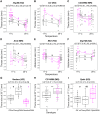
References
-
- Kingsolver JG, Huey RB. Size, temperature, and fitness: three rules. Evol Ecol Res. 2008;10:251–268.
-
- Peters RH. The ecological implications of body size. Cambridge: Cambridge University Press; 1986. 329 p.
-
- Smith FA, Lyons SK. Animal body size: linking pattern and process across space, time, and taxonomic group. University of Chicago Press; 2013. 267 p.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

