The structural basis of CstF-77 modulation of cleavage and polyadenylation through stimulation of CstF-64 activity
- PMID: 30257008
- PMCID: PMC6294498
- DOI: 10.1093/nar/gky862
The structural basis of CstF-77 modulation of cleavage and polyadenylation through stimulation of CstF-64 activity
Abstract
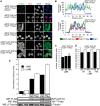
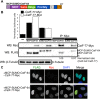
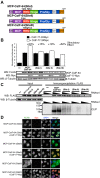
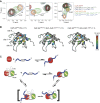
Cleavage and polyadenylation (C/P) of mRNA is an important cellular process that promotes increased diversity of mRNA isoforms and could change their stability in different cell types. The cleavage stimulation factor (CstF) complex, part of the C/P machinery, binds to U- and GU-rich sequences located downstream from the cleavage site through its RNA-binding subunit, CstF-64. Less is known about the function of the other two subunits of CstF, CstF-77 and CstF-50. Here, we show that the carboxy-terminus of CstF-77 plays a previously unrecognized role in enhancing C/P by altering how the RNA recognition motif (RRM) of CstF-64 binds RNA. In support of this finding, we also show that CstF-64 relies on CstF-77 to be transported to the nucleus; excess CstF-64 localizes to the cytoplasm, possibly via interaction with cytoplasmic RNAs. Reverse genetics and nuclear magnetic resonance studies of recombinant CstF-64 (RRM-Hinge) and CstF-77 (monkeytail-carboxy-terminal domain) indicate that the last 30 amino acids of CstF-77 increases the stability of the RRM, thus altering the affinity of the complex for RNA. These results provide new insights into the mechanism by which CstF regulates the location of the RNA cleavage site during C/P.
Figures



Similar articles
-
The hinge domain of the cleavage stimulation factor protein CstF-64 is essential for CstF-77 interaction, nuclear localization, and polyadenylation.J Biol Chem. 2010 Jan 1;285(1):695-704. doi: 10.1074/jbc.M109.061705. Epub 2009 Nov 3. J Biol Chem. 2010. PMID: 19887456 Free PMC article.
-
Cytoplasmic CstF-77 protein belongs to a masking complex with cytoplasmic polyadenylation element-binding protein in Xenopus oocytes.J Biol Chem. 2006 Sep 29;281(39):28687-98. doi: 10.1074/jbc.M601116200. Epub 2006 Aug 1. J Biol Chem. 2006. PMID: 16882666
-
The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location.Mol Cell Biol. 1994 Oct;14(10):6647-54. doi: 10.1128/mcb.14.10.6647-6654.1994. Mol Cell Biol. 1994. PMID: 7935383 Free PMC article.
-
The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5' capping and splicing.RNA Biol. 2011 Sep-Oct;8(5):748-53. doi: 10.4161/rna.8.5.16040. Epub 2011 Sep 1. RNA Biol. 2011. PMID: 21881408 Free PMC article. Review.
-
Recent molecular insights into canonical pre-mRNA 3'-end processing.Transcription. 2020 Apr;11(2):83-96. doi: 10.1080/21541264.2020.1777047. Epub 2020 Jun 11. Transcription. 2020. PMID: 32522085 Free PMC article. Review.
Cited by
-
Long noncoding RNA DLGAP1-AS2 promotes tumorigenesis and metastasis by regulating the Trim21/ELOA/LHPP axis in colorectal cancer.Mol Cancer. 2022 Nov 14;21(1):210. doi: 10.1186/s12943-022-01675-w. Mol Cancer. 2022. PMID: 36376892 Free PMC article.
-
Natural variation in the plant polyadenylation complex.Front Plant Sci. 2024 Jan 22;14:1303398. doi: 10.3389/fpls.2023.1303398. eCollection 2023. Front Plant Sci. 2024. PMID: 38317838 Free PMC article.
-
Plant terminators: the unsung heroes of gene expression.J Exp Bot. 2023 Apr 9;74(7):2239-2250. doi: 10.1093/jxb/erac467. J Exp Bot. 2023. PMID: 36477559 Free PMC article. Review.
-
Regulation of the Alternative Neural Transcriptome by ELAV/Hu RNA Binding Proteins.Front Genet. 2022 Feb 23;13:848626. doi: 10.3389/fgene.2022.848626. eCollection 2022. Front Genet. 2022. PMID: 35281806 Free PMC article. Review.
-
Electrostatic Interactions between CSTF2 and pre-mRNA Drive Cleavage and Polyadenylation.Biophys J. 2022 Feb 15;121(4):607-619. doi: 10.1016/j.bpj.2022.01.005. Epub 2022 Jan 26. Biophys J. 2022. PMID: 35090899 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

