Comparative genome analysis provides deep insights into Aeromonas hydrophila taxonomy and virulence-related factors
- PMID: 30257645
- PMCID: PMC6158803
- DOI: 10.1186/s12864-018-5100-4
Comparative genome analysis provides deep insights into Aeromonas hydrophila taxonomy and virulence-related factors
Abstract
Background: Aeromonas hydrophila is a potential zoonotic pathogen and primary fish pathogen. With overlapping characteristics, multiple isolates are often mislabelled and misclassified. Moreover, the potential pathogenic factors among the publicly available genomes in A. hydrophila strains of different origins have not yet been investigated.
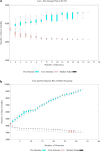
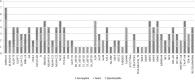
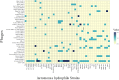
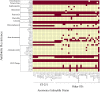
Results: To identify the valid strains of A. hydrophila and their pathogenic factors, we performed a pan-genomic study. It revealed that there were 13 mislabelled strains and 49 valid strains that were further verified by Average nucleotide identity (ANI), digital DNA-DNA hybridization (dDDH) and in silico multiple locus strain typing (MLST). Multiple numbers of phages were detected among the strains and among them Aeromonas phi 018 was frequently present. The diversity in type III secretion system (T3SS) and conservation of type II and type VI secretion systems (T2SS and T6SS, respectively) among all the strains are important to study for designing future strategies. The most prevalent antibiotic resistances were found to be beta-lactamase, polymyxin and colistin resistances. The comparative analyses of sequence type (ST) 251 and other ST groups revealed that there were higher numbers of virulence factors in ST-251 than in other STs group.
Conclusion: Publicly available genomes have 13 mislabelled organisms, and there are only 49 valid A. hydrophila strains. This valid pan-genome identifies multiple prophages that can be further utilized. Different A. hydrophila strains harbour multiple virulence factors and antibiotic resistance genes. Identification of such factors is important for designing future treatment regimes.
Keywords: Aeromonas; Antibiotic resistance patterns; Mislabelled strains; Pan-genome; Prophages; Virulence factors.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Stratev D, Vashin I, Rusev V. Prevalence and survival of Aeromonas spp. in foods - a review. Revue Med Vet. 2012;163:486–494.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

