Identification of Novel Natural Substrates of Fibroblast Activation Protein-alpha by Differential Degradomics and Proteomics
- PMID: 30257879
- PMCID: PMC6317473
- DOI: 10.1074/mcp.RA118.001046
Identification of Novel Natural Substrates of Fibroblast Activation Protein-alpha by Differential Degradomics and Proteomics
Abstract
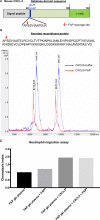
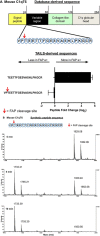
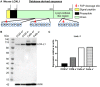
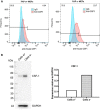
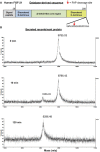
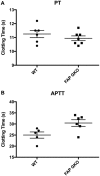
Fibroblast activation protein-alpha (FAP) is a cell-surface transmembrane-anchored dimeric protease. This unique, constitutively active serine protease has both dipeptidyl aminopeptidase and endopeptidase activities and can hydrolyze the post-proline bond. FAP expression is very low in adult organs but is upregulated by activated fibroblasts in sites of tissue remodeling, including fibrosis, atherosclerosis, arthritis and tumors. To identify the endogenous substrates of FAP, we immortalized primary mouse embryonic fibroblasts (MEFs) from FAP gene knockout embryos and then stably transduced them to express either enzymatically active or inactive FAP. The MEF secretomes were then analyzed using degradomic and proteomic techniques. Terminal amine isotopic labeling of substrates (TAILS)-based degradomics identified cleavage sites in collagens, many other extracellular matrix (ECM) and associated proteins, and lysyl oxidase-like-1, CXCL-5, CSF-1, and C1qT6, that were confirmed in vitro In addition, differential metabolic labeling coupled with quantitative proteomic analysis also implicated FAP in ECM-cell interactions, as well as with coagulation, metabolism and wound healing associated proteins. Plasma from FAP-deficient mice exhibited slower than wild-type clotting times. This study provides a significant expansion of the substrate repertoire of FAP and provides insight into the physiological and potential pathological roles of this enigmatic protease.
Keywords: Coagulation; Cytokines; Degradomics; Endopeptidases; Extracellular Matrix; Fibroblast Activation Protein; Fibroblasts; Proteases; SILAC; Substrate Identification.
© 2019 Zhang et al.
Conflict of interest statement
The authors declare no competing interests
Figures




References
-
- Park J. E., Lenter M. C., Zimmermann R. N., Garin-Chesa P., Old L. J., and Rettig W. J. (1999) Fibroblast activation protein: A dual-specificity serine protease expressed in reactive human tumor stromal fibroblasts. J. Biol. Chem. 274, 36505–36512 - PubMed
-
- Aertgeerts K., Levin I., Shi L., Snell G. P., Jennings A., Prasad G. S., Zhang Y., Kraus M. L., Salakian S., Sridhar V., Wijnands R., and Tennant M. G. (2005) Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein {alpha}. J. Biol. Chem. 280, 19441–19444 - PubMed
-
- Lee K. N., Jackson K. W., Christiansen V. J., Lee C. S., Chun J. G., and McKee P. A. (2006) Antiplasmin-cleaving enzyme is a soluble form of fibroblast activation protein. Blood 107, 1397–1404 - PubMed
-
- Hamson E. J., Keane F. M., Tholen S., Schilling O., and Gorrell M. D. (2014) Understanding Fibroblast Activation Protein (FAP): substrates, activities, expression and targeting for cancer therapy. Proteomics Clin. Appl. 8, 454–463 - PubMed
-
- Polak N., and Gorrell M. D. (2018) Fibroblast activation protein; FAP. In: Choi S., ed. Encyclopedia of Signaling Molecules, 2nd Ed., pp. 1676–1681, Springer International Publishing, Cham
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

