Role of transcription complexes in the formation of the basal methylation pattern in early development
- PMID: 30257947
- PMCID: PMC6187119
- DOI: 10.1073/pnas.1804755115
Role of transcription complexes in the formation of the basal methylation pattern in early development
Abstract
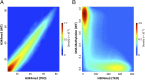
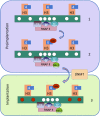
Following erasure in the blastocyst, the entire genome undergoes de novo methylation at the time of implantation, with CpG islands being protected from this process. This bimodal pattern is then preserved throughout development and the lifetime of the organism. Using mouse embryonic stem cells as a model system, we demonstrate that the binding of an RNA polymerase complex on DNA before de novo methylation is predictive of it being protected from this modification, and tethering experiments demonstrate that the presence of this complex is, in fact, sufficient to prevent methylation at these sites. This protection is most likely mediated by the recruitment of enzyme complexes that methylate histone H3K4 over a local region and, in this way, prevent access to the de novo methylation complex. The topological pattern of H3K4me3 that is formed while the DNA is as yet unmethylated provides a strikingly accurate template for modeling the genome-wide basal methylation pattern of the organism. These results have far-reaching consequences for understanding the relationship between RNA transcription and DNA methylation.
Keywords: development; epigenetics; histomodification; inheritance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. - PubMed
-
- Habibi E, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. - PubMed
-
- Brandeis M, et al. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

