Characterization of a Threonine-Rich Cluster in Hepatitis C Virus Nonstructural Protein 5A and Its Contribution to Hyperphosphorylation
- PMID: 30258001
- PMCID: PMC6258934
- DOI: 10.1128/JVI.00737-18
Characterization of a Threonine-Rich Cluster in Hepatitis C Virus Nonstructural Protein 5A and Its Contribution to Hyperphosphorylation
Abstract
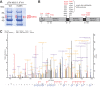
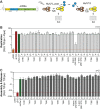
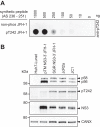
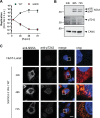
Hepatitis C virus (HCV) nonstructural protein 5A (NS5A) is a phosphoprotein with key functions in regulating viral RNA replication and assembly. Two phosphoisoforms are discriminated by their different apparent molecular weights: a basally phosphorylated (p56) and a hyperphosphorylated (p58) variant. The precise mechanisms governing p58 synthesis and specific functions of the isoforms are poorly understood. Our study aimed at a deeper understanding of determinants involved in p58 synthesis. We analyzed two variants of p56 and p58 of isolate JFH-1 separately by mass spectrometry using an expression model and thereby identified a threonine-rich phosphopeptide exclusively found in the hyperphosphorylated variant. Individual exchange of possible phosphoacceptor sites to phosphoablatant or -mimetic residues had little impact on HCV replication or assembly in cell culture. A phosphospecific antibody recognizing pT242 revealed that this position was indeed phosphorylated only in p58 and depended on casein kinase Iα. Importantly, phosphoablative mutations at positions T244 and S247 abrogated pT242 detection without substantial effects on global p58 levels, whereas mutations in the preceding serine-rich cluster dramatically reduced total p58 levels but had minor impact on pT242 levels, suggesting the existence of distinct subspecies of hyperphosphorylated NS5A. Mass spectrometry analyses of different genotypes showed variable phosphorylation patterns across NS5A and suggested that the threonine-rich region is also phosphorylated at T242 in gt4a and at S249 in gt1a, gt1b, and gt4a. Our data therefore indicate that p58 is not a single homogenously phosphorylated protein species but rather a population of various phosphoisoforms, with high variability between genotypes.IMPORTANCE Hepatitis C virus infections affect 71 million people worldwide and cause severe chronic liver disease. Recently, efficient antiviral therapies have been established, with inhibitors of nonstructural protein NS5A as a cornerstone. NS5A is a central regulator of HCV replication and assembly but is still enigmatic in its molecular functions. It exists in two phosphoisoforms, p56 and p58. We identified a phosphopeptide exclusively found in p58 and analyzed the determinants involved in phosphorylation of this region. We found evidence for very different phosphorylation patterns resulting in p58. These results challenge the concept of p58 being a homogenous species of NS5A molecules phosphorylated at the same positions and argues for at least two independently phosphorylated variants showing the same electrophoretic mobility, likely serving different functions.
Keywords: NS5A; PI4KA; PI4KIIIa; basal phosphorylation; hepatitis C virus; hyperphosphorylation; p56; p58; phosphorylation; positive-strand RNA virus.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
Serine 235 Is the Primary NS5A Hyperphosphorylation Site Responsible for Hepatitis C Virus Replication.J Virol. 2017 Jun 26;91(14):e00194-17. doi: 10.1128/JVI.00194-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28446668 Free PMC article.
-
Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-α in infectious virus production.J Virol. 2014 Jul;88(13):7541-55. doi: 10.1128/JVI.03170-13. Epub 2014 Apr 23. J Virol. 2014. PMID: 24760886 Free PMC article.
-
The alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation.J Virol. 2006 Nov;80(22):11305-12. doi: 10.1128/JVI.01465-06. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943283 Free PMC article.
-
Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication?Virology. 2007 Jul 20;364(1):1-9. doi: 10.1016/j.virol.2007.01.042. Epub 2007 Apr 2. Virology. 2007. PMID: 17400273 Review.
-
The non-structural 5A protein of hepatitis C virus.J Viral Hepat. 1999 Sep;6(5):343-56. doi: 10.1046/j.1365-2893.1999.00185.x. J Viral Hepat. 1999. PMID: 10607250 Review.
Cited by
-
Suppression of Innate Immunity by the Hepatitis C Virus (HCV): Revisiting the Specificity of Host-Virus Interactive Pathways.Int J Mol Sci. 2023 Nov 8;24(22):16100. doi: 10.3390/ijms242216100. Int J Mol Sci. 2023. PMID: 38003289 Free PMC article. Review.
-
Serine 229 Balances the Hepatitis C Virus Nonstructural Protein NS5A between Hypo- and Hyperphosphorylated States.J Virol. 2019 Nov 13;93(23):e01028-19. doi: 10.1128/JVI.01028-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31511391 Free PMC article.
-
Hepatitis C Virus Replication.Cold Spring Harb Perspect Med. 2020 Mar 2;10(3):a037093. doi: 10.1101/cshperspect.a037093. Cold Spring Harb Perspect Med. 2020. PMID: 31570388 Free PMC article. Review.
-
Phenotypic analysis of mutations at residue 146 provides insights into the relationship between NS5A hyperphosphorylation and hepatitis C virus genome replication.J Gen Virol. 2020 Mar;101(3):252-264. doi: 10.1099/jgv.0.001366. J Gen Virol. 2020. PMID: 31821131 Free PMC article.
-
Sequential Phosphorylation of the Hepatitis C Virus NS5A Protein Depends on NS3-Mediated Autocleavage between NS3 and NS4A.J Virol. 2020 Sep 15;94(19):e00420-20. doi: 10.1128/JVI.00420-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32699091 Free PMC article.
References
-
- Romero-Brey I, Berger C, Kallis S, Kolovou A, Paul D, Lohmann V, Bartenschlager R. 2015. NS5A domain 1 and polyprotein cleavage kinetics are critical for induction of double-membrane vesicles associated with hepatitis C virus replication. mBio 6:e00759. doi:10.1128/mBio.00759-15. - DOI - PMC - PubMed
-
- Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol 82:7964–7976. doi:10.1128/JVI.00826-08. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

