Novel Class of Viral Ankyrin Proteins Targeting the Host E3 Ubiquitin Ligase Cullin-2
- PMID: 30258003
- PMCID: PMC6232478
- DOI: 10.1128/JVI.01374-18
Novel Class of Viral Ankyrin Proteins Targeting the Host E3 Ubiquitin Ligase Cullin-2
Abstract
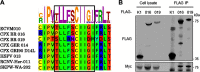
Ankyrin repeat (ANK) domains are among the most abundant motifs in eukaryotic proteins. ANK proteins are rare amongst viruses, with the exception of poxviruses, which presumably acquired them from the host via horizontal gene transfer. The architecture of poxvirus ANK proteins is, however, different from that of their cellular counterparts, and this precludes a direct acquisition event. Here we combine bioinformatics analysis and quantitative proteomics to discover a new class of viral ANK proteins with a domain organization that relates to cellular ANK proteins. These noncanonical viral ANK proteins, termed ANK/BC, interact with host Cullin-2 via a C-terminal BC box resembling that of cellular Cullin-2 substrate adaptors such as the von Hippel-Lindau protein. Mutagenesis of the BC box-like sequence abrogates binding to Cullin-2, whereas fusion of this motif to an ANK-only protein confers Cullin-2 association. We demonstrated that these viral ANK/BC proteins are potent immunomodulatory proteins suppressing the activation of the proinflammatory transcription factors NF-κB and interferon (IFN)-responsive factor 3 (IRF-3) and the production of cytokines and chemokines, including interferon, and that association with Cullin-2 is required for optimal inhibitory activity. ANK/BC proteins exist in several orthopoxviruses and cluster into 2 closely related orthologue groups in a phylogenetic lineage that is separate from that of canonical ANK/F-box proteins. Given the existence of cellular proteins with similar architecture, viral ANK/BC proteins may be closely related to the original ANK gene acquired by an ancestral orthopoxvirus. These findings uncover a novel viral strategy to antagonize innate immunity and shed light on the origin of the poxviral ANK protein family.IMPORTANCE Viruses encode multiple proteins aimed at modulating cellular homeostasis and antagonizing the host antiviral response. Most of these genes were originally acquired from the host and subsequently adapted to benefit the virus. ANK proteins are common in eukaryotes but are unusual amongst viruses, with the exception of poxviruses, where they represent one of the largest protein families. We report here the existence of a new class of viral ANK proteins, termed ANK/BC, that provide new insights into the origin of poxvirus ANK proteins. ANK/BC proteins target the host E3 ubiquitin ligase Cullin-2 via a C-terminal BC box domain and are potent suppressors of the production of inflammatory cytokines, including interferon. The existence of cellular ANK proteins whose architecture is similar suggests the acquisition of a host ANK/BC gene by an ancestral orthopoxvirus and its subsequent duplication and adaptation to widen the repertoire of immune evasion strategies.
Keywords: Ankyrin proteins; Cullin ubiquitin system; immune evasion; poxvirus.
Copyright © 2018 Odon et al.
Figures






Similar articles
-
Poxviral ankyrin proteins.Viruses. 2015 Feb 16;7(2):709-38. doi: 10.3390/v7020709. Viruses. 2015. PMID: 25690795 Free PMC article. Review.
-
F-box-like domains are present in most poxvirus ankyrin repeat proteins.Virus Genes. 2005 Oct;31(2):127-33. doi: 10.1007/s11262-005-1784-z. Virus Genes. 2005. PMID: 16025237
-
Chlorovirus Skp1-binding ankyrin repeat protein interplay and mimicry of cellular ubiquitin ligase machinery.J Virol. 2014 Dec;88(23):13798-810. doi: 10.1128/JVI.02109-14. Epub 2014 Sep 24. J Virol. 2014. PMID: 25253343 Free PMC article.
-
Vaccinia Virus BBK E3 Ligase Adaptor A55 Targets Importin-Dependent NF-κB Activation and Inhibits CD8+ T-Cell Memory.J Virol. 2019 May 1;93(10):e00051-19. doi: 10.1128/JVI.00051-19. Print 2019 May 15. J Virol. 2019. PMID: 30814284 Free PMC article.
-
Unique ankyrin repeat proteins in the genome of poxviruses-Boon or Wane, a critical review.Gene. 2024 Nov 15;927:148759. doi: 10.1016/j.gene.2024.148759. Epub 2024 Jul 9. Gene. 2024. PMID: 38992761 Review.
Cited by
-
Progress on Poxvirus E3 Ubiquitin Ligases and Adaptor Proteins.Front Immunol. 2021 Dec 9;12:740223. doi: 10.3389/fimmu.2021.740223. eCollection 2021. Front Immunol. 2021. PMID: 34956175 Free PMC article. Review.
-
Cullin-5 Adaptor SPSB1 Controls NF-κB Activation Downstream of Multiple Signaling Pathways.Front Immunol. 2020 Jan 21;10:3121. doi: 10.3389/fimmu.2019.03121. eCollection 2019. Front Immunol. 2020. PMID: 32038638 Free PMC article.
-
Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1.Viruses. 2024 Feb 3;16(2):247. doi: 10.3390/v16020247. Viruses. 2024. PMID: 38400023 Free PMC article.
-
Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS.Gut Microbes. 2020 Jul 3;11(4):771-788. doi: 10.1080/19490976.2019.1707015. Epub 2020 Jan 15. Gut Microbes. 2020. PMID: 31941397 Free PMC article.
-
Poxvirus Interactions with the Host Ubiquitin System.Pathogens. 2021 Aug 16;10(8):1034. doi: 10.3390/pathogens10081034. Pathogens. 2021. PMID: 34451498 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

