Lysosomal Proteases Are a Determinant of Coronavirus Tropism
- PMID: 30258004
- PMCID: PMC6258935
- DOI: 10.1128/JVI.01504-18
Lysosomal Proteases Are a Determinant of Coronavirus Tropism
Abstract
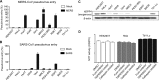
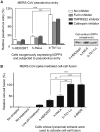
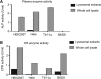
Cell entry by coronaviruses involves two principal steps, receptor binding and membrane fusion; the latter requires activation by host proteases, particularly lysosomal proteases. Despite the importance of lysosomal proteases in both coronavirus entry and cell metabolism, the correlation between lysosomal proteases and cell tropism of coronaviruses has not been established. Here, we examined the roles of lysosomal proteases in activating coronavirus surface spike proteins for membrane fusion, using the spike proteins from severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) as the model system. To this end, we controlled the contributions from receptor binding and other host proteases, thereby attributing coronavirus entry solely or mainly to the efficiency of lysosomal proteases in activating coronavirus spike-mediated membrane fusion. Our results showed that lysosomal proteases from bat cells support coronavirus spike-mediated pseudovirus entry and cell-cell fusion more effectively than their counterparts from human cells. Moreover, purified lysosomal extracts from bat cells cleave cell surface-expressed coronavirus spikes more efficiently than their counterparts from human cells. Overall, our study suggests that different lysosomal protease activities from different host species and tissue cells are an important determinant of the species and tissue tropism of coronaviruses.IMPORTANCE Coronaviruses are capable of colonizing new species, as evidenced by the recent emergence of SARS and MERS coronaviruses; they can also infect multiple tissues in the same species. Lysosomal proteases play critical roles in coronavirus entry by cleaving coronavirus surface spike proteins and activating the fusion of host and viral membranes; they also play critical roles in cell physiology by processing cellular products. How do different lysosomal protease activities from different cells impact coronavirus entry? Here, we controlled the contributions from known factors that function in coronavirus entry so that lysosomal protease activities became the only or the main determinant of coronavirus entry. Using pseudovirus entry, cell-cell fusion, and biochemical assays, we showed that lysosomal proteases from bat cells activate coronavirus spike-mediated membrane fusion more efficiently than their counterparts from human cells. Our study provides the first direct evidence supporting lysosomal proteases as a determinant of the species and tissue tropisms of coronaviruses.
Keywords: coronavirus spike protein; lysosomal proteases; species tropism; tissue tropism.
Copyright © 2018 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

