Spatiotemporal Distribution and Evolution of the A/H1N1 2009 Pandemic Influenza Virus in Pigs in France from 2009 to 2017: Identification of a Potential Swine-Specific Lineage
- PMID: 30258006
- PMCID: PMC6258933
- DOI: 10.1128/JVI.00988-18
Spatiotemporal Distribution and Evolution of the A/H1N1 2009 Pandemic Influenza Virus in Pigs in France from 2009 to 2017: Identification of a Potential Swine-Specific Lineage
Abstract
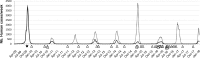
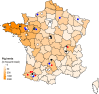
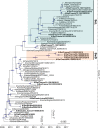
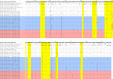
The H1N1 influenza virus responsible for the most recent pandemic in 2009 (H1N1pdm) has spread to swine populations worldwide while it replaced the previous seasonal H1N1 virus in humans. In France, surveillance of swine influenza A viruses in pig herds with respiratory outbreaks led to the detection of 44 H1N1pdm strains between 2009 and 2017, regardless of the season, and findings were not correlated with pig density. From these isolates, 17 whole-genome sequences were obtained, as were 6 additional hemagglutinin (HA)/neuraminidase (NA) sequences, in order to perform spatial and temporal analyses of genetic diversity and to compare evolutionary patterns of H1N1pdm in pigs to patterns for human strains. Following mutation accumulation and fixation over time, phylogenetic analyses revealed for the first time the divergence of a swine-specific genogroup within the H1N1pdm lineage. The divergence is thought to have occurred around 2011, although this was demonstrated only through strains isolated in 2015 to 2016 in the southern half of France. To date, these H1N1pdm swine strains have not been related to any increased virulence in swine herds and have not exhibited any antigenic drift compared to seasonal human strains. However, further monitoring is encouraged, as diverging evolutionary patterns in these two species, i.e., swine and humans, may lead to the emergence of viruses with a potentially higher risk to both animal and human health.IMPORTANCE Pigs are a "mixing vessel" for influenza A viruses (IAVs) because of their ability to be infected by avian and human IAVs and their propensity to facilitate viral genomic reassortment events. Also, as IAVs may evolve differently in swine and humans, pigs can become a reservoir for old human strains against which the human population has become immunologically naive. Thus, viruses from the novel swine-specific H1N1pdm genogroup may continue to diverge from seasonal H1N1pdm strains and/or from other H1N1pdm viruses infecting pigs and lead to the emergence of viruses that would not be covered by human vaccines and/or swine vaccines based on antigens closely related to the original H1N1pdm virus. This discovery confirms the importance of encouraging swine IAV monitoring because H1N1pdm swine viruses could carry an increased risk to both human and swine health in the future as a whole H1N1pdm virus or gene provider in subsequent reassortant viruses.
Keywords: antigenic drift; cat; genetic diversity; influenza A virus; molecular epidemiology; orthomyxoviridae; pandemic; pig; regional pattern; zoonotic infections.
Copyright © 2018 American Society for Microbiology.
Figures





References
-
- Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. - PMC - PubMed
-
- Centers for Disease Control and Prevention, NCIRD. 2017. Influenza type A viruses. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/flu/avianflu/influenza-a-virus-subtypes.htm.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

