Biofilm Formation Drives Transfer of the Conjugative Element ICE Bs1 in Bacillus subtilis
- PMID: 30258041
- PMCID: PMC6158512
- DOI: 10.1128/mSphere.00473-18
Biofilm Formation Drives Transfer of the Conjugative Element ICE Bs1 in Bacillus subtilis
Abstract
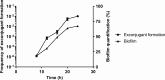
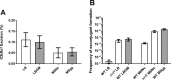
Horizontal gene transfer by integrative and conjugative elements (ICEs) is a very important mechanism for spreading antibiotic resistance in various bacterial species. In environmental and clinical settings, most bacteria form biofilms as a way to protect themselves against extracellular stress. However, much remains to be known about ICE transfer in biofilms. Using ICEBs1 from Bacillus subtilis, we show that the natural conjugation efficiency of this ICE is greatly affected by the ability of the donor and recipient to form a biofilm. ICEBs1 transfer considerably increases in biofilm, even at low donor/recipient ratios. Also, while there is a clear temporal correlation between biofilm formation and ICEBs1 transfer, biofilms do not alter the level of ICEBs1 excision in donor cells. Conjugative transfer appears to be favored by the biophysical context of biofilms. Indeed, extracellular matrix production, particularly from the recipient cells, is essential for biofilms to promote ICEBs1 transfer. Our study provides basic new knowledge on the high rate of conjugative transfer of ICEs in biofilms, a widely preponderant bacterial lifestyle in the environment, which could have a major impact on our understanding of horizontal gene transfer in natural and clinical environments.IMPORTANCE Transfer of mobile genetic elements from one bacterium to another is the principal cause of the spread of antibiotic resistance. However, the dissemination of these elements in environmental contexts is poorly understood. In clinical and environmental settings, bacteria are often found living in multicellular communities encased in a matrix, a structure known as a biofilm. In this study, we examined how forming a biofilm influences the transmission of an integrative and conjugative element (ICE). Using the model Gram-positive bacterium B. subtilis, we observed that biofilm formation highly favors ICE transfer. This increase in conjugative transfer is due to the production of extracellular matrix, which creates an ideal biophysical context. Our study provides important insights into the role of the biofilm structure in driving conjugative transfer, which is of major importance since biofilm is a widely preponderant bacterial lifestyle for clinically relevant bacterial strains.
Keywords: Bacillus subtilis; ICEBs1; biofilms; extracellular matrix; horizontal gene transfer.
Copyright © 2018 Lécuyer et al.
Figures



References
-
- Moreno Switt AI, den Bakker HC, Cummings CA, Rodriguez-Rivera LD, Govoni G, Raneiri ML, Degoricija L, Brown S, Hoelzer K, Peters JE, Bolchacova E, Furtado MR, Wiedmann M. 2012. Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One 7:e41247. doi: 10.1371/journal.pone.0041247. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

