UBL3 modification influences protein sorting to small extracellular vesicles
- PMID: 30258067
- PMCID: PMC6158211
- DOI: 10.1038/s41467-018-06197-y
UBL3 modification influences protein sorting to small extracellular vesicles
Abstract
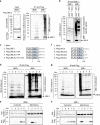
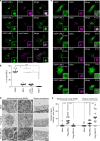
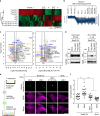
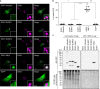
Exosomes, a type of small extracellular vesicles (sEVs), derived from multivesicular bodies (MVBs), mediate cell-to-cell communication by transporting proteins, mRNAs, and miRNAs. However, the molecular mechanism by which proteins are sorted to sEVs is not fully understood. Here, we report that ubiquitin-like 3 (UBL3)/membrane-anchored Ub-fold protein (MUB) acts as a posttranslational modification (PTM) factor that regulates protein sorting to sEVs. We find that UBL3 modification is indispensable for sorting of UBL3 to MVBs and sEVs. We also observe a 60% reduction of total protein levels in sEVs purified from Ubl3-knockout mice compared with those from wild-type mice. By performing proteomics analysis, we find 1241 UBL3-interacting proteins, including Ras. We also show that UBL3 directly modifies Ras and oncogenic RasG12V mutant, and that UBL3 expression enhances sorting of RasG12V to sEVs via UBL3 modification. Collectively, these results indicate that PTM by UBL3 influences the sorting of proteins to sEVs.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

