Meta-analysis of data from spaceflight transcriptome experiments does not support the idea of a common bacterial "spaceflight response"
- PMID: 30258082
- PMCID: PMC6158273
- DOI: 10.1038/s41598-018-32818-z
Meta-analysis of data from spaceflight transcriptome experiments does not support the idea of a common bacterial "spaceflight response"
Abstract
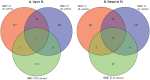
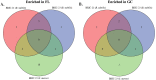
Several studies have been undertaken with the goal of understanding how bacterial transcriptomes respond to the human spaceflight environment. However, these experiments have been conducted using a variety of organisms, media, culture conditions, and spaceflight hardware, and to date no cross-experiment analyses have been performed to uncover possible commonalities in their responses. In this study, eight bacterial transcriptome datasets deposited in NASA's GeneLab Data System were standardized through a common bioinformatics pipeline then subjected to meta-analysis to identify among the datasets (i) individual genes which might be significantly differentially expressed, or (ii) gene sets which might be significantly enriched. Neither analysis resulted in identification of responses shared among all datasets. Principal Component Analysis of the data revealed that most of the variation in the datasets derived from differences in the experiments themselves.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Comparisons of Transcriptome Profiles from Bacillus subtilis Cells Grown in Space versus High Aspect Ratio Vessel (HARV) Clinostats Reveal a Low Degree of Concordance.Astrobiology. 2020 Dec;20(12):1498-1509. doi: 10.1089/ast.2020.2235. Epub 2020 Oct 19. Astrobiology. 2020. PMID: 33074712
-
Test of Arabidopsis Space Transcriptome: A Discovery Environment to Explore Multiple Plant Biology Spaceflight Experiments.Front Plant Sci. 2020 Mar 4;11:147. doi: 10.3389/fpls.2020.00147. eCollection 2020. Front Plant Sci. 2020. PMID: 32265943 Free PMC article.
-
Global transcriptomic analysis suggests carbon dioxide as an environmental stressor in spaceflight: A systems biology GeneLab case study.Sci Rep. 2018 Mar 8;8(1):4191. doi: 10.1038/s41598-018-22613-1. Sci Rep. 2018. PMID: 29520055 Free PMC article.
-
Conducting Plant Experiments in Space and on the Moon.Methods Mol Biol. 2022;2368:165-198. doi: 10.1007/978-1-0716-1677-2_12. Methods Mol Biol. 2022. PMID: 34647256 Review.
-
Spaceflight bioreactor studies of cells and tissues.Adv Space Biol Med. 2002;8:177-95. doi: 10.1016/s1569-2574(02)08019-x. Adv Space Biol Med. 2002. PMID: 12951697 Review.
Cited by
-
CAMDLES: CFD-DEM Simulation of Microbial Communities in Spaceflight and Artificial Microgravity.Life (Basel). 2022 Apr 29;12(5):660. doi: 10.3390/life12050660. Life (Basel). 2022. PMID: 35629329 Free PMC article.
-
Clinostat Rotation Affects Metabolite Transportation and Increases Organic Acid Production by Aspergillus carbonarius, as Revealed by Differential Metabolomic Analysis.Appl Environ Microbiol. 2019 Aug 29;85(18):e01023-19. doi: 10.1128/AEM.01023-19. Print 2019 Sep 15. Appl Environ Microbiol. 2019. PMID: 31300399 Free PMC article.
-
One Step Forward with Dry Surface Biofilm (DSB) of Staphylococcus aureus: TMT-Based Quantitative Proteomic Analysis Reveals Proteomic Shifts between DSB and Hydrated Biofilm.Int J Mol Sci. 2022 Oct 13;23(20):12238. doi: 10.3390/ijms232012238. Int J Mol Sci. 2022. PMID: 36293092 Free PMC article.
-
Microgravity and evasion of plant innate immunity by human bacterial pathogens.NPJ Microgravity. 2023 Sep 7;9(1):71. doi: 10.1038/s41526-023-00323-x. NPJ Microgravity. 2023. PMID: 37679341 Free PMC article. Review.
-
Mechanotransduction in Prokaryotes: A Possible Mechanism of Spaceflight Adaptation.Life (Basel). 2021 Jan 7;11(1):33. doi: 10.3390/life11010033. Life (Basel). 2021. PMID: 33430182 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

