The Molecular Basis for Specificity at the Level of the Protein Kinase a Catalytic Subunit
- PMID: 30258407
- PMCID: PMC6143667
- DOI: 10.3389/fendo.2018.00538
The Molecular Basis for Specificity at the Level of the Protein Kinase a Catalytic Subunit
Abstract
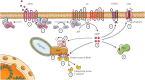
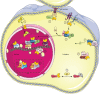
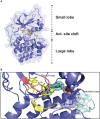
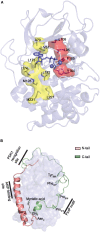
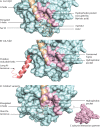
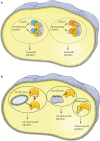
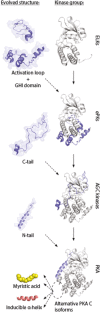
Assembly of multi enzyme complexes at subcellular localizations by anchoring- and scaffolding proteins represents a pivotal mechanism for achieving spatiotemporal regulation of cellular signaling after hormone receptor targeting [for review, see (1)]. In the 3' 5'-cyclic adenosine monophosphate (cAMP) dependent protein kinase (PKA) signaling pathway it is generally accepted that specificity is secured at several levels. This includes at the first level stimulation of receptors coupled to heterotrimeric G proteins which through stimulation of adenylyl cyclase (AC) forms the second messenger cAMP. Cyclic AMP has several receptors including PKA. PKA is a tetrameric holoenzyme consisting of a regulatory (R) subunit dimer and two catalytic (C) subunits. The R subunit is the receptor for cAMP and compartmentalizes cAMP signals through binding to cell and tissue-specifically expressed A kinase anchoring proteins (AKAPs). The current dogma tells that in the presence of cAMP, PKA dissociates into an R subunit dimer and two C subunits which are free to phosphorylate relevant substrates in the cytosol and nucleus. The release of the C subunit has raised the question how specificity of the cAMP and PKA signaling pathway is maintained when the C subunit no longer is attached to the R subunit-AKAP complex. An increasing body of evidence points toward a regulatory role of the cAMP and PKA signaling pathway by targeting the C subunits to various C subunit binding proteins in the cytosol and nucleus. Moreover, recent identification of isoform specific amino acid sequences, motifs and three dimensional structures have together provided new insight into how PKA at the level of the C subunit may act in a highly isoform-specific fashion. Here we discuss recent understanding of specificity of the cAMP and PKA signaling pathway based on C subunit subcellular targeting as well as evolution of the C subunit structure that may contribute to the dynamic regulation of C subunit activity.
Keywords: PKA; anchoring; catalytic subunit; molecular determinants; specificity.
Figures
References
-
- Skålhegg BS, Taskén K. Specificity in the cAMP/PKA signaling pathway. Differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci. (2000) 5:678–93. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

