Traditional Norwegian Kveik Are a Genetically Distinct Group of Domesticated Saccharomyces cerevisiae Brewing Yeasts
- PMID: 30258422
- PMCID: PMC6145013
- DOI: 10.3389/fmicb.2018.02137
Traditional Norwegian Kveik Are a Genetically Distinct Group of Domesticated Saccharomyces cerevisiae Brewing Yeasts
Abstract
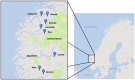
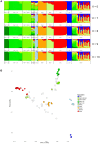
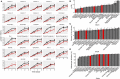
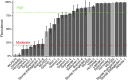
The widespread production of fermented food and beverages has resulted in the domestication of Saccharomyces cerevisiae yeasts specifically adapted to beer production. While there is evidence beer yeast domestication was accelerated by industrialization of beer, there also exists a farmhouse brewing culture in western Norway which has passed down yeasts referred to as kveik for generations. This practice has resulted in ale yeasts which are typically highly flocculant, phenolic off flavor negative (POF-), and exhibit a high rate of fermentation, similar to previously characterized lineages of domesticated yeast. Additionally, kveik yeasts are reportedly high-temperature tolerant, likely due to the traditional practice of pitching yeast into warm (>28°C) wort. Here, we characterize kveik yeasts from 9 different Norwegian sources via PCR fingerprinting, whole genome sequencing of selected strains, phenotypic screens, and lab-scale fermentations. Phylogenetic analysis suggests that kveik yeasts form a distinct group among beer yeasts. Additionally, we identify a novel POF- loss-of-function mutation, as well as SNPs and CNVs potentially relevant to the thermotolerance, high ethanol tolerance, and high fermentation rate phenotypes of kveik strains. We also identify domestication markers related to flocculation in kveik. Taken together, the results suggest that Norwegian kveik yeasts are a genetically distinct group of domesticated beer yeasts with properties highly relevant to the brewing sector.
Keywords: Saccharomyces; ale; brewing; domestication; fermentation; kveik; yeast.
Figures

Similar articles
-
Kveik Brewing Yeasts Demonstrate Wide Flexibility in Beer Fermentation Temperature Tolerance and Exhibit Enhanced Trehalose Accumulation.Front Microbiol. 2022 Mar 16;13:747546. doi: 10.3389/fmicb.2022.747546. eCollection 2022. Front Microbiol. 2022. PMID: 35369501 Free PMC article.
-
Co-Fermentations of Kveik with Non-Conventional Yeasts for Targeted Aroma Modulation.Microorganisms. 2022 Sep 27;10(10):1922. doi: 10.3390/microorganisms10101922. Microorganisms. 2022. PMID: 36296198 Free PMC article.
-
European farmhouse brewing yeasts form a distinct genetic group.Appl Microbiol Biotechnol. 2024 Aug 2;108(1):430. doi: 10.1007/s00253-024-13267-3. Appl Microbiol Biotechnol. 2024. PMID: 39093468 Free PMC article.
-
[Non-conventional yeasts as tools for innovation and differentiation in brewing].Rev Argent Microbiol. 2021 Oct-Dec;53(4):359-377. doi: 10.1016/j.ram.2021.01.003. Epub 2021 Mar 3. Rev Argent Microbiol. 2021. PMID: 33674169 Review. Spanish.
-
Adaptive Laboratory Evolution of Ale and Lager Yeasts for Improved Brewing Efficiency and Beer Quality.Annu Rev Food Sci Technol. 2020 Mar 25;11:23-44. doi: 10.1146/annurev-food-032519-051715. Epub 2020 Jan 17. Annu Rev Food Sci Technol. 2020. PMID: 31951488 Review.
Cited by
-
A Quasi-Domesticate Relic Hybrid Population of Saccharomyces cerevisiae × S. paradoxus Adapted to Olive Brine.Front Genet. 2019 May 29;10:449. doi: 10.3389/fgene.2019.00449. eCollection 2019. Front Genet. 2019. PMID: 31191600 Free PMC article.
-
Never Change a Brewing Yeast? Why Not, There Are Plenty to Choose From.Front Genet. 2020 Nov 6;11:582789. doi: 10.3389/fgene.2020.582789. eCollection 2020. Front Genet. 2020. PMID: 33240329 Free PMC article. Review.
-
A Unique Saccharomyces cerevisiae × Saccharomyces uvarum Hybrid Isolated From Norwegian Farmhouse Beer: Characterization and Reconstruction.Front Microbiol. 2018 Sep 24;9:2253. doi: 10.3389/fmicb.2018.02253. eCollection 2018. Front Microbiol. 2018. PMID: 30319573 Free PMC article.
-
Sour Beer as Bioreservoir of Novel Craft Ale Yeast Cultures.Microorganisms. 2023 Aug 23;11(9):2138. doi: 10.3390/microorganisms11092138. Microorganisms. 2023. PMID: 37763982 Free PMC article.
-
Isolation and characterization of haploid heterothallic beer yeasts.Appl Microbiol Biotechnol. 2025 Jan 22;109(1):17. doi: 10.1007/s00253-024-13397-8. Appl Microbiol Biotechnol. 2025. PMID: 39841271 Free PMC article.
References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Institute.
-
- ASBC (2011). Yeast−11. ASBC Methods of Analysis, 1–2.
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (2002). Short Protocols in Molecular Biology, 5th Edn. Hoboken, NJ: Wiley.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

