Ratiometric Raman imaging reveals the new anti-cancer potential of lipid targeting drugs
- PMID: 30258563
- PMCID: PMC6128370
- DOI: 10.1039/c8sc02312c
Ratiometric Raman imaging reveals the new anti-cancer potential of lipid targeting drugs
Abstract
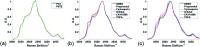
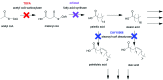
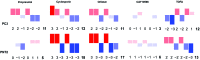
De novo lipid synthesis is upregulated in cancer cells and inhibiting these pathways has displayed anti-tumour activity. Here we use Raman spectroscopy, focusing solely on high wavenumber spectra, to detect changes in lipid composition in single cells in response to drugs targeting de novo lipid synthesis. Unexpectedly, the beta-blocker propranolol showed selectively towards cancerous PC3 compared to non-cancerous PNT2 prostate cells, demonstrating the potential of this approach to identify new anti-cancer drug leads. A unique and simple ratiometric approach for intracellular lipid investigation is reported using statistical analysis to create phenotypic 'barcodes', a globally applicable strategy for Raman drug-cell studies. High wavenumber spectral analysis is compatible with low cost glass substrates, easily translatable into the cytological work stream. The analytical strength of this technique could have a significant impact on cancer treatment through vastly improved understanding of cancer cell metabolism, and thus guide drug design and enhance personalised medicine strategies.
Figures

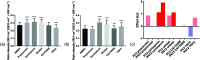
References
-
- Siegel R. L., Miller K. D., Jemal A. Ca-Cancer J. Clin. 2016;66:7–30. - PubMed
-
- Tennant D. A., Duran R. V., Gottlieb E. Nat. Rev. Cancer. 2010;10:267–277. - PubMed
-
- Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. - PubMed
-
- Koppenol W. H., Bounds P. L., Dang C. V. Nat. Rev. Cancer. 2011;11:325–337. - PubMed
-
- Warburg O. Science. 1956;123:309–314. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

