Metabolite-Centric Reporter Pathway and Tripartite Network Analysis of Arabidopsi s Under Cold Stress
- PMID: 30258841
- PMCID: PMC6143811
- DOI: 10.3389/fbioe.2018.00121
Metabolite-Centric Reporter Pathway and Tripartite Network Analysis of Arabidopsi s Under Cold Stress
Abstract
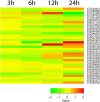
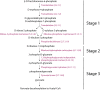
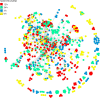
The study of plant resistance to cold stress and the metabolic processes underlying its molecular mechanisms benefit crop improvement programs. Here we investigate the effects of cold stress on the metabolic pathways of Arabidopsis when directly inferred at system level from transcriptome data. A metabolite-centric reporter pathway analysis approach enabled the computation of metabolites associated with transcripts at four time points of cold treatment. Tripartite networks of gene-metabolite-pathway connectivity outlined the response of metabolites and pathways to cold stress. Our metabolome-independent analysis revealed stress-associated metabolites in pathway routes of the cold stress response, including amino acid, carbohydrate, lipid, hormone, energy, photosynthesis, and signaling pathways. Cold stress first triggered the mobilization of energy from glycolysis and ethanol degradation to enhance TCA cycle activity via acetyl-CoA. Interestingly, tripartite networks lacked power law behavior and scale free connectivity, favoring modularity. Network rewiring explicitly involved energetics, signal, carbon and redox metabolisms and membrane remodeling.
Keywords: cold stress; microarray; network modularity; pathway analysis; power law; reporter metabolite; reporter pathway.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources

