InfiniumPurify: An R package for estimating and accounting for tumor purity in cancer methylation research
- PMID: 30258934
- PMCID: PMC6147081
- DOI: 10.1016/j.gendis.2018.02.003
InfiniumPurify: An R package for estimating and accounting for tumor purity in cancer methylation research
Abstract
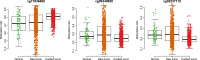
The proposition of cancer cells in a tumor sample, named as tumor purity, is an intrinsic factor of tumor samples and has potentially great influence in variety of analyses including differential methylation, subclonal deconvolution and subtype clustering. InfiniumPurify is an integrated R package for estimating and accounting for tumor purity based on DNA methylation Infinium 450 k array data. InfiniumPurify has three main functions getPurity, InfiniumDMC and InfiniumClust, which could infer tumor purity, differential methylation analysis and tumor sample cluster accounting for estimated or user-provided tumor purities, respectively. The InfiniumPurify package provides a comprehensive analysis of tumor purity in cancer methylation research.
Keywords: Cancer subtype classification; DNA methylation; Differential methylation analysis; InfiniumPurify; Tumor purity.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

