ACTB and SDHA Are Suitable Endogenous Reference Genes for Gene Expression Studies in Human Astrocytomas Using Quantitative RT-PCR
- PMID: 30259794
- PMCID: PMC6161201
- DOI: 10.1177/1533033818802318
ACTB and SDHA Are Suitable Endogenous Reference Genes for Gene Expression Studies in Human Astrocytomas Using Quantitative RT-PCR
Abstract
Background: Quantitative real-time reverse-transcription polymerase chain reaction is frequently used as research tool in experimental oncology. There are some studies of valid endogenous control genes in the field of human glioma research, which, however, only focus on the comparison between normal brain with tumor tissue and malignant transformation toward secondary glioblastomas. Aim of this study was to validate a more general reference gene also suitable for pre- and posttreatment analysis and other evaluations (eg, primary vs secondary glioblastoma).
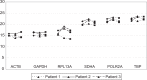
Methods: This quantitative polymerase chain reaction analysis was performed to test a panel of the 6 most suitable reference genes from other studies representing different physiological pathways (ACTB, GAPDH, POLR2A, RPL13A, SDHA, and TBP) in all common glioma groups, namely: diffuse astrocytoma World Health Organization II, anaplastic astrocytoma World Health Organization III, secondary glioblastoma World Health Organization IV with and without chemotherapy, primary glioblastoma, recurrent glioblastoma, and gliomas before and after radiation. Expression stability was tested during the longitudinal course of the disease in 8 single patients.
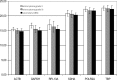
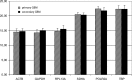
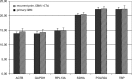
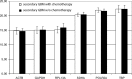
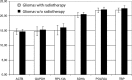
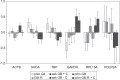
Results: Evaluation of the expression levels of the 6 target genes showed that ACTB, GAPDH, and RPL13A show higher expression compared to SDHA, POLR2A, and TBP. ACTB, GAPDH, and RPL13A showed different expression levels between astrozytoma grade II and primary glioblastoma. Except for this difference, the candidate genes were not differentially expressed between primary and secondary glioblastomas and between the World Health Organization tumor grades. Furthermore, they remained stable before and after radiotherapy and/or chemotherapy. Therefore, they are adequate references for glioblastoma gene expression studies. The comparison of all tested genes resulted in SDHA and ACTB as most stable reference genes determined by the NormFinder software. Our data revealed lowest intragroup variation in the SDHA, highest in the RPL13A gene.
Conclusions: All tested genes may be recommended as universal reference genes for data normalization in gene expression studies under different treatment regimens both in primary glioblastomas and astrocytomas of different grades (World Health Organization grades II-IV), respectively. In summary, ACTB and SDHA exhibited the best stability values and showed the lowest intergroup expression variability.
Keywords: WHO grade; astrocytoma; glioblastoma multiforme; glioma; housekeeping gene; qPCR; quantitative RT-PCR; quantitative real-time polymerase chain reaction; reference gene.
Conflict of interest statement
Figures

Similar articles
-
Identification of valid endogenous control genes for determining gene expression in human glioma.Neuro Oncol. 2010 Jun;12(6):570-9. doi: 10.1093/neuonc/nop072. Epub 2010 Feb 5. Neuro Oncol. 2010. PMID: 20511187 Free PMC article.
-
Selection of suitable reference genes for expression analysis in human glioma using RT-qPCR.J Neurooncol. 2015 May;123(1):35-42. doi: 10.1007/s11060-015-1772-7. Epub 2015 Apr 11. J Neurooncol. 2015. PMID: 25862007
-
Identification of endogenous reference genes for qRT-PCR analysis in normal matched breast tumor tissues.Oncol Res. 2009;17(8):353-65. doi: 10.3727/096504009788428460. Oncol Res. 2009. PMID: 19544972
-
Molecular biology of malignant gliomas.Clin Transl Oncol. 2006 Sep;8(9):635-41. doi: 10.1007/s12094-006-0033-9. Clin Transl Oncol. 2006. PMID: 17005465 Review.
-
[Gemistocytic astrocytomas].Arkh Patol. 2018;80(4):27-38. doi: 10.17116/patol201880427. Arkh Patol. 2018. PMID: 30059069 Russian.
Cited by
-
Selection of Reliable Reference Genes for Analysis of Gene Expression in Spinal Cord during Rat Postnatal Development and after Injury.Brain Sci. 2019 Dec 20;10(1):6. doi: 10.3390/brainsci10010006. Brain Sci. 2019. PMID: 31861889 Free PMC article.
-
Identification and Verification of Key Genes Associated with Temozolomide Resistance in Glioblastoma Based on Comprehensive Bioinformatics Analysis.Iran J Biotechnol. 2024 Oct 1;22(4):e3892. doi: 10.30498/ijb.2024.448826.3892. eCollection 2024 Oct. Iran J Biotechnol. 2024. PMID: 40225293 Free PMC article.
-
Molecular and Histological Evaluation of Sheep Ovarian Tissue Subjected to Lyophilization.Animals (Basel). 2021 Nov 29;11(12):3407. doi: 10.3390/ani11123407. Animals (Basel). 2021. PMID: 34944182 Free PMC article.
-
Selection of Stable Reference Genes for Gene Expression Studies in Activated and Non-Activated PBMCs Under Normoxic and Hypoxic Conditions.Int J Mol Sci. 2025 Jul 15;26(14):6790. doi: 10.3390/ijms26146790. Int J Mol Sci. 2025. PMID: 40725038 Free PMC article.
-
Low RNA stability signifies strong expression regulatability of tumor suppressors.Nucleic Acids Res. 2023 Nov 27;51(21):11534-11548. doi: 10.1093/nar/gkad838. Nucleic Acids Res. 2023. PMID: 37831104 Free PMC article.
References
-
- Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. - PubMed
-
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol, 2016: 131(6)803–20 - PubMed
-
- Malzkorn B, Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr Opin Oncol, 2016: 28(6)494–501. - PubMed
-
- Mishima K, Kato Y, Kaneko MK, Nishikawa R, Hirose T, Matsutani M. Increased expression of podoplanin in malignant astrocytic tumors as a novel molecular marker of malignant progression. Acta Neuropathol. 2006;111(5):483–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

