Recent progress in enzymatic protein labelling techniques and their applications
- PMID: 30259933
- PMCID: PMC6289631
- DOI: 10.1039/c8cs00537k
Recent progress in enzymatic protein labelling techniques and their applications
Abstract
Protein-based conjugates are valuable constructs for a variety of applications. Conjugation of proteins to fluorophores is commonly used to study their cellular localization and the protein-protein interactions. Modification of therapeutic proteins with either polymers or cytotoxic moieties greatly enhances their pharmacokinetics or potency. To label a protein of interest, conventional direct chemical reaction with the side-chains of native amino acids often yields heterogeneously modified products. This renders their characterization complicated, requires difficult separation steps and may impact protein function. Although modification can also be achieved via the insertion of unnatural amino acids bearing bioorthogonal functional groups, these methods can have lower protein expression yields, limiting large scale production. As a site-specific modification method, enzymatic protein labelling is highly efficient and robust under mild reaction conditions. Significant progress has been made over the last five years in modifying proteins using enzymatic methods for numerous applications, including the creation of clinically relevant conjugates with polymers, cytotoxins or imaging agents, fluorescent or affinity probes to study complex protein interaction networks, and protein-linked materials for biosensing. This review summarizes developments in enzymatic protein labelling over the last five years for a panel of ten enzymes, including sortase A, subtiligase, microbial transglutaminase, farnesyltransferase, N-myristoyltransferase, phosphopantetheinyl transferases, tubulin tyrosin ligase, lipoic acid ligase, biotin ligase and formylglycine generating enzyme.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare
Figures
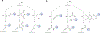





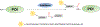


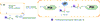
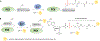



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

